* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download Chapter 8
Survey
Document related concepts
History of genetic engineering wikipedia , lookup
Medical genetics wikipedia , lookup
Dual inheritance theory wikipedia , lookup
Deoxyribozyme wikipedia , lookup
Dominance (genetics) wikipedia , lookup
Designer baby wikipedia , lookup
Human genetic variation wikipedia , lookup
Behavioural genetics wikipedia , lookup
Genetic drift wikipedia , lookup
Polymorphism (biology) wikipedia , lookup
Quantitative trait locus wikipedia , lookup
Microevolution wikipedia , lookup
Population genetics wikipedia , lookup
Natural selection wikipedia , lookup
Transcript
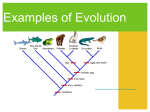
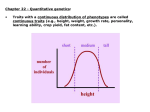
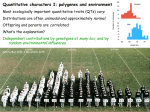
Chapter 8 Quantitative Genetics Polygenic Inheritance: when a number of different pairs of alleles at several loci are important for expression of a trait. Such traits are typically quantitative in nature, not qualitative. Quantitative Genetics: study of traits that show continuous variation and are due to the combined effects of many different loci, as well as the environment. Some Quantitative Traits in Humans Fig 8.1 Mendelian Genetics can Explain Quantitative Traits Figure 8.2 East’s Data confirming the Predictions of Mendelian Genetics Fig 8.3 What is the cause of this phenotypic variation in height? For an individual: Phenotype = genotype + environment P=G+E For a population, measure phenotypic variance: VP = VG + VE Heritability: the fraction of the total variance in a trait that is due to variance explained by genotypes. In the broad sense (using clones, full-sibs) heritability = VG/VP = VG/(VG + VE) Estimating Heritability from Parents and Offspring Offspring do not Offspring moderately Offspring strongly resemble parents resemble parents resemble parents Figure 8.11 -Scatterplots showing offspring height as a function of parent height The slope of these lines represents a version of heritability called narrow-sense heritability or h2 Partitioning Genetic Effects Additive effects (VA): effects where the contribution of an allele to the phenotype is independent of identity of other alleles at the same or at different loci. Dominance (VD): non-additive interactions of alleles at one locus Epistasis (VI): non-additive interactions of alleles at different loci Maternal effects(VM): due to mother’s environment and her ability to provision resources for offspring. Additive Genetic Variance versus Dominance Figure 8.13 Epistasis The total genetic variation for a population is the sum of these effects: VG = VA + VD + VI + VM • Genetic environment does not alter additive effects, so additive effects are the basis for a response to selection • Non-additive genetic variance is dependent on the alleles at the same loci and alleles occurring at other loci narrow-sense heritability = h2 = VA/VP with VP = VA + VD + VI + VM + VE Example 1 Fig 8.11d Example 2: Heritability of beak size in song sparrows Measuring Natural Selection (differences in survival and reproductive success) Fig 8.15 Measuring Natural Selection cont. Selection differential (S): the difference in mean phenotype between the selected individuals (survivors/breeders) and the entire population. Selection gradient: slope of the line showing relative fitness as a function of phenotype. selection gradient = the selection differential/ population variance Selection gradient: • Assign absolute fitnesses • Convert absolute fitnesses to relative fitnesses • Make a scatterplot of relative fitness and calculate the slope of the line of best fit Determining the Response to Selection Response to selection (R): the difference between the mean phenotype of the offspring when selection takes place and the mean phenotype of the offspring if selection does not take place. Response to Selection cont. Response to Selection cont. h2 = the slope of the line = rise/run = mean of selected offspring – mean of all offspring ___________________________________________ mean of selected parents – mean of all parents h2 = R/S Response to Selection cont. Therefore, R = h2 * S In other words, the strength of the response to selection depends upon the relative contribution of additive genetic variance to the phenotype and the strength of natural selection. Revisiting Modes of Selection Directional selection Fitness consistently increases or decreases with the value of a trait Changes the mean value of the trait; potentially reduces the variance. Stabilizing selection Individuals with intermediate values of a trait have higher fitness Does not change the mean; reduces the variance. Disruptive selection Individuals with extreme values of a trait have the higher fitness Does not change the mean; increases the variance. Does not necessarily cause a bimodal distribution. Revisiting Modes of Selection cont. Following 7 years of growth: Heritability for flower size was between 0.2 and 1.0. Slope = 0.13. Selection differential = gradient * variance. 0.13 * 5.66 = 0.74 mm. 0.74/14.2 = 5% change if flower size for selection differential Response to selection was 9% Concerns about the Environment Heritability is dependent upon the population being examined, as well as the environment that is being experienced. Environment can alter both the mean of a trait, the variance, and how heritable the trait is. Recall: h2 = VG/(VG + VE). So, the only way to determine the cause of differences between populations is to rear individuals from each of the populations in identical environments. Clausen, Keck and Hiesey’s (1948) work with the perennial wild flower, Achillea High heritability within populations tells us nothing about the cause of differences between populations What does this suggest about Human IQ and the Book, The Bell Curve Fallacy? (Murray and Herrstein, 1994) Clausen, Keck and Hiesey’s (1948) work with the perennial wild flower, Achillea THE END. Genetic Constraints on Evolution through N. Selection: Remember, requirements for a response to selection: (1) Natural selection (diff. survival/reproduction) (2) Phenotypic variation in a given trait (3) Underlying genetic variation Constraints: (1) Absence of genetic variation (2) Correlated Characters: Pleiotropic effects: effects of single genes on multiple traits Linkage: the tendency for alleles at different loci to be inherited together. Selection will favor linkage dis-equilibrium if certain combinations of alleles have greater fitness than other combinations. Example: Adaptive landscape of two traits for garter snakes (striped versus spotted and straight-line escape versus reversals). Striped, straight-line runners and spotted, reversals are more fit than the other two combinations. Recombination can constrain natural selection in this case.