* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download doc
Survey
Document related concepts
Protein moonlighting wikipedia , lookup
Biochemical switches in the cell cycle wikipedia , lookup
Protein phosphorylation wikipedia , lookup
Hedgehog signaling pathway wikipedia , lookup
G protein–coupled receptor wikipedia , lookup
Cellular differentiation wikipedia , lookup
Purinergic signalling wikipedia , lookup
List of types of proteins wikipedia , lookup
Histone acetylation and deacetylation wikipedia , lookup
Mitogen-activated protein kinase wikipedia , lookup
Paracrine signalling wikipedia , lookup
Transcript
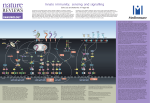
Part 1. Signal transduction leading to NF-B activation. NF-B is a dimeric transcription factor composed of five proteins (RelA, p50, c-Rel, p52 and RelB) maintained inactive in the cytoplasm by inhibitory proteins (IBs). Upon activation, NF-B is released from the inhibitory proteins and translocate to the cell nucleus where it transactivates target genes. There are two distinct NF-B-activation pathways: the classical and the alternative. The classical pathway was the first being characterized and is triggered by pro-inflammatory signals such as cytokines, bacterial and viral infections, all of which activate the IKK complex. This complex is composed of two catalytic subunits, IKK and IKK and a regulatory subunit IKK (also known as NEMO). This complex phosphorylates NF-B bound IBs, thereby targeting them for proteasomal degradation and liberating NF-B dimers to enter the nucleus and mediate transcription of target genes. This reaction mostly depends on the catalytic subunit IKK which carries out IB phosphorylation. The alternative NF-B activation pathway which is triggered by some ligands from the TNF family (such as BAFF, LT, CD40L,….) involves the upstream kinase NF-B-inducing kinase (NIK) activating IKK homodimers, independently of either IKK or IKK. phosphorylation and processing of p100. This leads to the The two pathways switch on different gene sets and therefore mediate different immune functions. The contribution of the classical pathway to acute inflammation and cell survival mechanisms is well accepted, and sustained NF-B in various malignancies has been described. Owing to the variety of target genes activated by the classical pathway, which include those encoding cytokines, chemokines, proteases and inhibitors of apoptosis, it has been proposed that the classical NF-B activation pathway might also link inflammation to tumour promotion and progression. Therefore, an emerging link between chronic inflammation and cancer is being established and some cancers can also be considered as chronic inflammatory diseases. In this part of the proposal, we will better characterize and compare the two signal transduction pathways leading to NF-B activation, as well as their crosstalk with some other signalling pathways that contribute to proinflammatory gene expression. In addition, we will address several molecular mechanisms of signal attenuation that determine the kinetics and strength of NF-B activation. Regulation of NF-B-dependent gene expression will not only be studied at the level of the initiating and propagating signalling events in the cytoplasm, but also at the level of the gene in the nucleus. WP1: Study of the classical and alternative pathways of NF-B activation in cellular models. Although the knowledge of NF-B activation in response to various receptors has increased considerably and has uncovered a crucial role for protein-protein interactions, multiple aspects remain unclear. We will analyse the potential role of specific signalling molecules at the cross-road of NF-B and other signalling pathways (e.g the IRF pathway) that determine a proper immune response. Special attention will also be given to the molecular mechanisms that regulate the multiple proteinprotein interactions in NF-B signalling in response to various stimuli (e.g. TNF, TLRs and TCR). In this context, we will study the role of protein ubiquitination and acetylation. WP1.1 Characterization of signalling molecules at the cross-road of NF-B and other signalling pathways Molecular mechanisms of IRF and NF-B activation through TANK ULG2 studies the scaffold protein TANK/I-TRAF, which is a TRAF2 and IKK-interacting protein that is required for the TNF-induced and NF-B-dependent expression of a selection of target genes (Bonif et al., 2006). The fact that TANK also binds TBK1 and IKK kinases, which are essential components of the IRF signalling pathway (Fitzgerald et al., 2003), suggests also a role for TANK in IRF activation. This will be further addressed by deciphering the phase of NF-B and IRF activation in TANK-deficient macrophages treated with various stimuli, by identifying TANK-dependent genes in LPS-treated macrophages through micro-array analyses, and by functionally characterizing the role of novel TANKinteracting proteins (e.g. IRF7) that were recently identified by yeast two-hybrid screening (ULG2, unpublished data). Moreover the role of specific posttranslational modifications of TANK (phosphorylation, ubiquitination) will also be studied by ULG2 and UG2. Caspase-mediated activation of NF-B The proteolytic activity of caspases mainly connotes their central role in apoptosis and inflammation. UG2 and others recently discovered that caspase-1, -2 and -8 can also activate NF-B through the recruitment of specific NF-B-signalling molecules in different caspase containing protein complexes: the inflammasome (caspase-1 and caspase-8) or the stressosome (caspase-2) (reviewed in Lamkanfi et. al., 2006). In this project, the formation of the stressosome under different conditions of cellular stress (DNA damage, ATP release, ER stress, mitochondrial ROS, heat, TLR) will be studied by UG2 in cells that are deficient in different signalling molecules, using gel filtration, blue native electrophoresis, confocal microscopy, immunoprecipitation and mass spectrometry analysis (see also WP6.2). Structure-function analysis of the CARD domains of caspase-1 and caspase-2 will reveal the potential of CARD-only proteins to interfere with NF-B activation in macrophages (UG2 and ULG1). Modulation of NOD2-dependent NF-B signalling by the actin cytoskeleton The cytosolic Nod2 protein plays a key role in sensing bacteria and generating an inflammatory immune response that is mediated by the activation of NF-B (reviewed by Strober et al., 2006). Specific mutations in the Nod2 gene have been associated with Crohn disease (CD). To date, little information exists about the regulation of the Nod2-signalling pathway. ULG1 recently showed that Nod2 is recruited in membrane ruffles through Rac1 and that actin disruption significantly increased Nod2-mediated NF-B activation (unpublished data). The recruitment of Nod2 in dynamic cytoskeletal structures could be a strategy to both repress Nod2-dependent NF-B signalling in unstimulated cells and rapidly mobilize Nod2 during infection with invasive pathogens that interfere with the cytoskeleton machinery. ULG1 will investigate this further in a model of apical invasion of Nod2-transfected polarized intestinal epithelial cells by Salmonella. Nod2 recruitment to Salmonella invasion sites and interactions of Nod2 with proteins involved in actin reorganization (e.g. Rac1) will be examined by colocalization (confocal microscopy) and coimmunoprecipitation assays. The role of Salmonellainduced reorganisation of the cytoskeleton machinery will be analysed by ULG1 and UG2 using specific Salmonella type III secretion mutants or cells expressing specific Rac1 mutants. ULG1 and UG2 will also try to determine the domains and/or residues of Nod2 involved in its recruitment to invasion sites as well as the behaviour of CD-associated Nod2 mutants. Role of Protein kinase D in NF-B signalling pathways KUL in collaboration with UG2 aims to investigate the contribution of Protein Kinase D (PKD) in signalling towards NF-B in response to Reactive Oxygen Species (ROS), TLR or TCR stimulation. ROS leads to Abl-mediated tyrosine phosphorylation of PKD, which is both necessary and sufficient to activate NF-B (Storz and Toker, 2003). KUL recently found that tyrosine phosphorylated PKD induces an invasive phenotype in normal and malignant cells (unpublished data) and will investigate in collaboration with UG2 the underlying molecular mechanisms of NF-B activation by tyrosine phosphorylated PKD, as well as the role of NF-B activation in the invasion promoting effect of PKD. Furthermore, KUL and UG2 will investigate whether these effects can be regulated by ABINs and whether other tyrosine kinases (besides Abl) can phosphorylate PKD. PKD is a signalling target of PKC and the latter plays a crucial role in TLR4-induced NF-B activation (Castrillo et al., 2001). KUL and UG2 will therefore investigate whether TLR4 activates PKD, whether this activation occurs through PKC and whether PKD is required for TLR4-induced NF-B activation. Since PKD has been shown to associate with the kinase Btk and since Btk plays a crucial role in TLRinduced NF-B activation (Jefferies et al., 2003), KUL and UG2 will also investigate the potential role for a PKD-Btk complex in TLR-induced NF-B activation. A role for PKD-mediated activation and phosphorylation of HPK1 in TCR-induced NF-B activation has recently been demonstrated (Arnold et al., 2005). Since PKD is a downstream target of PKC-, KUL and UG2 want to investigate whether PKD is part of the PKC-/CARMA/Bcl10/MALT induced NF-B activation pathway in response to TCR. Molecular crosstalk between oxysteroid nuclear receptors and NF-B in foamy macrophages M. tuberculosis infects and survives within macrophages, the cells that are out to provide an effective initial barrier to contain infection. Mycolic acid (MA), a characteristic biolipid of the cell wall of M. tuberculosis was recently discovered by UG1 to mimic pathogen-associated host innate immune response (Korf et al., 2005). A major feature of the MA-induced macrophage response is a disruption of lipid homeostasis, resulting in excessive cholesterol accumulation and the development of foamy macrophages. This imbalance between cholesterol uptake and export was reflected also at the molecular level as apparent from the increased mRNA levels of CD36, involved in the uptake of oxidized LDL, and the decreased levels of the cholesterol transporter, ABCA1, and its extracellular acceptor, APOE. Finally, a striking induction of the cholesterol-sensing oxysteroid nuclear receptor, LXR, was observed. LXR and other lipid-activated transcription factors such as PPAR, functioning as lipid sensors and guarding the lipid homeostasis within the cell, have also been implicated in the control of innate immune response genes (Joseph et al., 2004). In agreement herewith, MA-treated macrophages exhibit a markedly altered responsiveness to LPS, increased expression of TNF and IDO, decreased expression of IL-10, and novel expression of IFN- and MPO. Within the present proposal, UG1 intends to define in more details the molecular pathways through which accumulation of cholesterol modifies innate immune functions of MA-induced foamy macrophages. TLR4 signalling pathways leading to NF-B and IRF3/STAT1 activation will be verified in macrophages treated in culture with MA under conditions of normal and low oxidized LDL, thus allowing verifying the interference of cholesterol accumulation. The cross-talk between oxysteroid receptors and NF-B will be verified by using macrophages isolated from mouse strains deficient for the A and b LXR paralogues and/or treated with LXR and PPRA agonists. Also the response pattern of macrophages deficient for TANK, a scaffold protein required for the NF-B dependent expression of a subset of target genes will be investigated by UG1 and ULG2. Regulation of IKK complex activity through protein acetylation. In addition to protein ubiquitination (Chen, 2005), protein acetylation has emerged as a key regulatory element in the field of NF-B-dependent gene expression (Quivy and Van Lint, 2004). ULG1 has demonstrated a strong synergistic transcriptional activation of NF-B-dependent promoters (IL-6, HIV1, ICAM-1, IL-8) by NF-B inducers of the classical pathway (TNF, IL-1, PMA and pervanadate) and deacetylase (HDAC) inhibitors. Mechanistically, HDAC inhibitors prolonged the activated IKK complex activity. This prolongation could explain the persistent proteasome-mediated degradation of the inhibitory protein IB and the resulting prolonged presence and DNA-binding of NF-B in the nucleus (Quivy et al., 2002; Adam et al., 2003; Quivy and Van Lint, 2004). These results suggest that HDAC(s) control(s) directly or indirectly the activity of the IKK complex. In this regard, we have shown that several HDACs interact with the IKK complex probably via IKK. Within the current proposal, ULG1 will compare, in normal versus HDAC knock down cell lines, the activation kinetics of the NF-B pathway and their functional consequences. Interestingly with pervanadate, in contrast to the other NF-B inducers, ULG1 observed no prolongation of IKK activation by HDAC inhibitors, but a downregulation of the IB mRNA transcription, and UG1, ULG1 will examine the molecular basis for this difference. WP1.2. Study of the alternative pathway of NF-B activation in cellular models. In our studies on the alternative NF-B activation pathway, ULG1 will mainly focus on NF-B activation in response to the Lymphotoxin- Receptor (LTR) (Dejardin et al., 2002). Biochemical and genetic studies have demonstrated a role for this receptor in the development, as well as in the maintenance of the architecture, of secondary lymphoid organs. Abnormal spatio-temporal expression of the LTR is associated with the formation of ectopic lymphoid structures, which is a hallmark of most chronic inflammatory diseases. So far, the only kinases known to play a role in the activation of the alternative pathway are NIK and IKK but the mechanisms controlling their kinase activity is not well defined (Pomerantz and Baltimore, 2002). Phosphorylation and dephosphorylation of signalling proteins allow to turn on and/or off most signalling pathways in eukaryotic cells and have been studied for decades. Although the phosphoacceptor sites within the kinase domain of NIK and IKK that modulate their kinase activity have been identified, it is still not known whether they are the only requirement for the full activation of the enzymatic activities of NIK and IKK mediating the processing of p100. There is a growing body of evidence that ubiquitination and de-ubiquitination activities tightly control a wide range of signalling pathways, TNF signalling being one striking example (reviewed by Chen, 2002). Whereas the mechanism of A20-mediated de-ubiquitination of polyubiquitinated RIP via K63-linkage followed by the polyubiquitination of RIP via K48-linkage is well defined for the negative feedback regulation of TNF-induced NF-B signalling (reviewed by Heyninck and Beyaert, 2005), up to now no such mechanism has been discovered for the alternative NF-B pathway. ULG1, EU1 & UG2 will investigate if and how monoubiquitination and/or the various ubiquitin linkages of polyubiquitination reactions control positively and/or negatively the activation of the alternative pathway. Another goal is to understand how TRAF proteins control the induction of the alternative pathway. Indeed, preliminary data indicate that TRAF2 and TRAF3 have an inhibitory function on the alternative NF-B pathway as opposed to their positive role in the classical NF-B pathway. How the inhibitory function of TRAF proteins is alleviated upon ligation of the LTR is still an unanswered question. First of all, post-translational modifications of TRAF proteins that occur following the engagement of the LTR will be investigated by ULG1 and UG2. In this regard, the putative role of the protein TANK/ITRAF will be studied. Indeed, it has been shown that TANK exclusively binds TRAF2 and TRAF3. The putative role of c-IAP1 for the steady state of TRAF2 and TRAF3 will also be investigated by ULG1 and ULG2. Finally, a forward genetic approach based on the use of a chemical mutagenic compound to identify new proteins specific to the LTR-mediated alternative NF-B pathway will be conducted by ULG1, ULG2, and EU1. A particular attention will be given to new IKK-interacting partners (EU1) via an in vivo biotinylation approach (De Boer et al., 2003). Potential new candidates will be targeted with siRNA or lentivirus-expressing shRNA to downregulate their level of expression both in vitro system and in vivo. The effect of the RNA interference will be evaluated by ULG1 in vivo in an adjuvant arthritis model in rats for which it would be feasible to inject the siRNA or lentirus-expressing shRNA directly into the joints. WP2. Signal modulation of NF-B activation An effective immune system requires rapid and appropriate activation of an inflammatory response upon infection, in order to terminate the spread of the infection as quickly as possible. However, as the inflammatory response evolved at the cost of self-tissue damage, failure of resolution can have detrimental effects to the organism and can result in the development of chronic inflammatory disorders. Because of this inherent danger of inflammation, the crucial activation steps of this process are tightly regulated. Hence, many cellular proteins control the NF-B-signalling pathways induced by pathogens and pro-inflammatory cytokines. Moreover, also microbes themselves have developed several strategies to modulate NF-B and IRF mediated signalling and evade the immune system. These checkpoints act at different levels in NF-B signalling and apply various strategies to inhibit NFB activation, such as interfering with protein-protein interactions, ubiquitination and acetylation. Understanding these negative regulatory mechanisms of NF-B activation is of great value for the development of new approaches to treat NF-B-mediated inflammatory diseases. WP2.1. Mechanisms of signal attenuation of NF-B activation in the cytoplasm. UG2 aims to study the negative feedback regulation of NF-B activation by A20 and ABINs (respectively ABIN-1, -2 and –3). They previously showed that ABINs can inhibit NF-B activation upstream of IKK in response to various stimuli in vitro as well as in murine models of hepatitis and asthma (e.g. Wullaert et al., 2005; El Bakkouri et al., 2005). To further elucidate their mechanism of action as well as the mechanisms that regulate their activity, A20 and ABINs will be subjected to detailed protein-protein interaction studies. Yeast two-hybrid screening already revealed some interesting candidates that are currently analysed (UG2 & ULG2). UG2 will also identify novel A20and ABIN-interacting proteins via ‘Mammalian Protein-Protein Interaction Trapping’ (MAPPIT) (Eyckerman et al., 2001), as well as via TAP-tagged affinity purification of an ‘in vivo’ expressed TAPA20 transgene (see also WP5). The role of A20, ABINs and their interacting proteins will be functionally analysed using RNAi as well as cells derived from knockout mice. Conditional ABIN-1 and –3 knockout mice will therefore be generated by UG2 during this project (see also WP5). Interesting perspectives are also gained from the recent demonstration that A20 has dual ubiquitin ligase and deubiquitinase activities (reviewed by Heyninck and Beyaert, 2005). Using complementation studies of A20-deficient MEF cells with specific A20 mutants, UG2 will investigate the specific role of this dual ubiquitin-editing activity of A20 in the regulation of specific ubiquitin-dependent protein-protein interactions that are involved in NF-B and IRF activation. In addition, UG2 will identify and characterize several known and novel A20-binding proteins as potential substrates or regulators of A20 activity. UG2 studies the regulation of NF-B activation by MyD88s, which they previously identified as an alternative splice variant of the IL-1R and TLR-adaptor protein MyD88 that is produced in LPS-treated monocytes (Janssens et al., 2002). MyD88s prevents IL-1 and TLR induced NF-B activation by inhibiting the recruitment of IRAK4 to the receptor complex. The effect of MyD88s will now also be analysed on other MyD88-dependent and MyD88-independent signalling pathways, including the IRF pathway (in collaboration with ULG2). UG2 has clear evidence that TNF leads to a serious and irreversible inhibition of ligand binding of the glucocorticoid receptor (GR) and that this forms the basis of excessive NF-B activity (unpublished data). In the context of this project, UG2 will try to identify the mechanism of GR inhibition as well as its protection by HSP70 (Van Molle et al., 2002). Therefore, they will analyse GR modifications in response to TNF, thereby focusing on the role of IKKs, MAP kinases and MKPs (UG1 and UG2). Therefore, UG1 and UG2 will analyse GR modifications in response to TNF, thereby focusing on the role of IKKs, MAP kinases and MKPs. WP2.2. Interference of pathogens with NF-B and IRF signalling pathways. ULG1 showed that Varicella-zoster virus (VZV) can activate NF-B and IRF3 via TLR-dependent and independent signalling, but that infected cells become resistant to TNF and to IFN- (unpublished data). In this project ULG1 will try to identify the viral components and the intracellular signalling components involved in the activation of NF-B and IRF3 through TLR2 and TLR3, and investigate the potential role of intracellular receptors such as RIG1 or PKR. In addition, we will study the putative role of two viral kinases in NF-B and IRF3 activation, as well as the molecular basis for the inducible resistance to TNF and IFN-. The latter will be addressed by detailed analysis of the signalling pathways in infected cells and by microarray analysis of the genes modulated by VZV infection (in collaboration with UG2 and ULG2). WP3: Nuclear mechanisms in NF-B signalling. Once NF-B has migrated into the nucleus, it transactivates a large variety of target promoters with a gene specificity and an activation strength and duration, which is ensured by post-translational modifications (including phosphorylation and acetylation), by the availability of co-acting transcription factors and non-DNA-bound cofactors, and by the recruitment of chromatin-modifying enzymatic complexes. WP3.1. Characterization of cofactor complexes involved in NF-B-driven gene expression UG1 and others have demonstrated crucial involvement of multiple kinases (i.e. PKAc, RSK1/2, MSK1/2, and IKK) to fine-tune NF-B-dependent gene expression at the transcription factor, cofactor (SMRT, CBP) and chromatin (histone H3) level (Vermeulen et al., 2003). However, the interplay with other cofactor families [histone acetyltransferases (HATs), histone deacetylases (HDACs), histone methyltransferases (HMTs)] is less well understood. UG1, EU1 and EU2 will examine time-dependent cofactor associations, localization and nucleocytoplasmic shuttling dynamics during gene activation (via classical or alternative pathways) or gene repression by glucocorticoids and/or PPAR nuclear receptor agonists by means of immunoaffinity and/or immunofluorescence approaches. Functional involvement of individual or tandem cofactor complexes in gene expression/repression of IL-6 gene will be further evaluated by siRNA tools or KO cells. Co-immunoprecipitation assays of tagged cofactors in the presence of absence of kinase inhibitors, deacetylase inhibitors (HDACs), or (de)methylase inhibitors should allow dissecting relationships in co-factor complex formation and/or activities in a gene-specific way. WP3.2. Role of epigenetic modifications in IL-6 transcriptional regulation. IL-6 is a tightly controlled inflammatory cytokine which preserves immune homeostasis, whereas excessive IL-6 amounts causes chronic inflammatory disorders and tumorigenesis. Various stimuli, like oxidative stress, viruses (HIV, HTLV), DNA damage, classical (TNFR, TLR, IL1R) and alternative (CD40, BAFF, LT) NF-B signalling pathways, can trigger IL-6 gene expression. Previous studies have focused on IL-6 promoter chromatin regulation (nucleosome mapping, chromatin accessibility assays) in benign and metastatic breast cancer cells, which differentially express the IL-6 gene (Ndlovu et al., 2006). Within the current proposal, UG1 will further analyse IL-6 promoter DNA methylation patterns in presence or absence of DNA methylation inhibitors (5’-azacytidine) or PARP inhibitors (i.e. aminobenzamide, which has been described to elicit DNA methylation) in relation to chromatin accessibility in breast cancer cells (MDA-MB231 vs. MCF7), epithelial cells (A549, TC10) and immune cells (primary B cells, dendritic cells) in response to different stimuli. Toll-like receptors (TLRs), which activate innate and adaptive immune responses, are thought to be restricted to immune cells. However, TLRs are also (over)expressed on (metastatic) breast tumor cells, suggesting that TLR activation may be an important event in tumor cell immune evasion. UG1 will further analyse in breast cancer cells the effect of epigenetic cancer drugs (5’-azacytidine, HDAC inhibitors, PARP inhibitors) on the “inflammatory” epigenetic network (DNA methylation, histone modifications, nucleosome remodelling) and IL-6 gene expression modifications in response to endogenously generated (HSPs, HMBG1) or exogenous (LPS) stimuli and/or TLR antagonists (siRNA, E5564, peptides) WP3.3. Combinatorial control: NF-B activation and other activation cascades. Bacterial toxins were found to interfere with pathogen responses via MAPK and NF-B signalling pathways. This allowed the identification of a subclass of MAPK-sensitive NF-B target genes with combinatorial promoter regulation by CREB and NF-B. UG1 will focus on the role of MSK/RSK kinases in mediating gene-specific crosstalk between CREB and NF-B (dimerization, localization, modifications, chromatin accessibility) at the IL-6 gene promoter. Another example of combinatorial control concerns differential gene repression efficacy of glucocorticoids on NF-B (De Bosscher et al., 2003) versus NF-B-IRF3 target genes in response to TLR signalling. UG1 will focus on promoterspecific GR-NF-B versus GR-NF-B/IRF3 transrepression mechanisms during inflammatory gene responses. As PPAR negatively interferes with inflammatory gene expression through interference with NF-B (Delerive et al., 1999), UG1 and EU2 will additionally explore promoter-specific PPAR-NFB versus PPAR-NF-B/IRF3 transrepression mechanisms and interference between PPAR and GR. WP3.4. Implication of Sir2/Sirt1 in NF-kB-driven gene expression associated with aging. As aging is associated with accumulation of DNA damage and with increased IL-6 expression levels, various strategies are aimed to protect against inflammation syndrome-related aging discomforts. Interestingly, caloric restriction or caloric mimetics (polyphenolic compounds) protect against aging via regulation of Sir2/Sirt1 (a class III HDAC). Important targets of Sirt1 are p53, NF-B and Ku70. UG1 and ULG1 will investigate IL-6 gene regulation by p53, NF-B and Ku70 in response to DNA damage pathways (ATM/PIDD) in wild-type and Sir2 KO cells. Furthermore, UG1 and ULG1 will investigate concomitant acetylation of selected factors which may affect cofactor interaction and/or subcellular localization.