* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download Huang, David, Center for Structural Biochemistry
Protein (nutrient) wikipedia , lookup
Biochemistry wikipedia , lookup
Protein moonlighting wikipedia , lookup
Protein folding wikipedia , lookup
Drug design wikipedia , lookup
G protein–coupled receptor wikipedia , lookup
Protein domain wikipedia , lookup
List of types of proteins wikipedia , lookup
Metalloprotein wikipedia , lookup
Homology modeling wikipedia , lookup
Interactome wikipedia , lookup
Western blot wikipedia , lookup
Two-hybrid screening wikipedia , lookup
Protein adsorption wikipedia , lookup
X-ray crystallography wikipedia , lookup
Proteolysis wikipedia , lookup
Intrinsically disordered proteins wikipedia , lookup
Protein purification wikipedia , lookup
Protein structure prediction wikipedia , lookup
Protein–protein interaction wikipedia , lookup
Nuclear magnetic resonance spectroscopy of proteins wikipedia , lookup
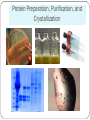
Center for Structural Biochemistry Montpellier, France David Huang 2015 Protein Preparation, Purification, and Crystallization Preparation – Plasmids were transformed into competent cells and the specific protein was expressed and collected. Purification – Protein was purified using a nickel affinity column and a gel filtration column. Crystallization – Purified proteins were prepared with various solvent and ligands on crystallography plates. Crystals were collected after a few days. Protein Preparation, Purification, and Crystallization Data Collection, Structure Determination, and Analysis Data Collection – Electron density data were collected from the protein crystals using x-ray crystallography. Structure Determination – The electron density data was used to determine the structure of the proteins in complex with the ligands using the COOT software. Analysis – The specific hydrogen bonds or hydrophobic interactions around the ligand binding pocket were analyzed after the structure had been determined. Data Collection, Structure Determination, and Analysis Most Rewarding Aspect of My Work Experience I was able to travel to Grenoble to use the ESRF (European Synchroton Radiation Facility), a large particle accelerator that allows us to collect well resolved x-ray diffraction data. Collecting good x-ray diffraction data and solving these structures was the culmination of all the weeks I spent in the lab doing bench work preparing proteins and crystals. European Synchrotron Radiation Facility How My Work Impacted the Organization I helped to solve the structures of PXR (pregnane X receptor) in complex with the ligands oxadiazon, pretilachlore, fipronil, fenvalerate, αzearalenol, 4-tert-octylphenol and 2 other unnamed synthetic ligands. Solving these structures give us a better understanding of how the protein binds to different ligands and will help us with structure based rational drug design. Ligands Pretilachlore Fenvalerate Oxadiazon α-Zearalenol Fipronil 4-tert-octylphenol Academic Choices and Career Plans This internship helped me solidify my interest in doing research and made me certain that I want to continue doing research in graduate school. Since I still prefer chemistry research over structural biochemistry research, I hope to work in the relevant field of synthesizing ligands and drug molecules based on the structural data of proteins. Cultural Awareness/Personal Growth Beyond Montpellier, I traveled to Sete, St.Guilhem, Grenoble, Paris, and Barcelona and experienced French and Spanish culture through food and travel. I met a lot of international students and learned their perspective on American politics and education. I learned to be truly independent by surviving in a country that spoke a foreign language, working at a 9 to 6 job everyday, and cooking all meals for myself. Travel Travel Travel Acknowledgments Princeton and IIP Luisa Duarte-Silva Centre de Biochimie Structurale Dr. William Bourguet Dr. Vanessa Delfosse Dr. Deborah Harrus