* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download nCounter® Virtual Cell Cycle Gene Set
Biology and consumer behaviour wikipedia , lookup
Neuronal ceroid lipofuscinosis wikipedia , lookup
Epigenetics of diabetes Type 2 wikipedia , lookup
Ridge (biology) wikipedia , lookup
Genetic engineering wikipedia , lookup
X-inactivation wikipedia , lookup
Genome evolution wikipedia , lookup
Genomic imprinting wikipedia , lookup
Minimal genome wikipedia , lookup
Epigenetics in stem-cell differentiation wikipedia , lookup
History of genetic engineering wikipedia , lookup
Nutriepigenomics wikipedia , lookup
Gene desert wikipedia , lookup
Gene therapy wikipedia , lookup
Gene nomenclature wikipedia , lookup
Polycomb Group Proteins and Cancer wikipedia , lookup
Gene expression programming wikipedia , lookup
Genome (book) wikipedia , lookup
Gene therapy of the human retina wikipedia , lookup
Therapeutic gene modulation wikipedia , lookup
Microevolution wikipedia , lookup
Epigenetics of human development wikipedia , lookup
Site-specific recombinase technology wikipedia , lookup
Mir-92 microRNA precursor family wikipedia , lookup
Gene expression profiling wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Vectors in gene therapy wikipedia , lookup
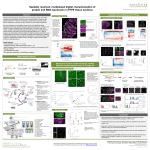
Product Data Sheet nCounter® Virtual Cell Cycle Gene Set Product Highlights •Simple No more days spent cross referencing databases •Highly curated Our expert bioinformaticists use a very rigorous process to select the most meaningful set of genes •Efficient Multiplexed assay profiles 183 cell cycle-related genes in one reaction •Cost-effective Gold-standard data at a fraction of the cost A Gene Set You Can Count On The nCounter Virtual Cell Cycle Gene Set is a comprehensive list of 183 human genes known to be differentially expressed in various phases of the cell cycle. The gene list spans a broad range of cell cycle-related biological processes, including: • • • • • cell proliferation and differentiation transcriptional regulation oncogenesis tumor suppression DNA metabolism, replication and repair processes The nCounter Virtual Cell Cycle Gene Set, takes the guesswork out of gene selection allowing researchers to accelerate their research and quickly generate expression data for a large panel of cell cycle-related genes. The gene list was compiled by querying several public databases for cell cycle-related genes. This list was refined using multiple criteria, including scoring each gene for relevance in cell cycle-related pathways using IPA (by Ingenuity® Systems, Inc). Each gene was also verified to be differentially expressed in various System Performance Critical Specifications Level of Multiplexing Recommended Amount of Starting Material Sample Types Supported Reaction Volume Limit of Detection Fold Change Sensitivity Spike Correlation Linear Dynamic Range Controls 183 genes known to be differentially expressed in cell cycle 100ng of total RNA, or lysate from ~10,000 cells Total RNA, Cell Lysates in GITC, FFPE derived total RNA and PAXgene lysed whole blood, Amplified RNA 30 µL 0.5fM spike-in control (~1 copy per cell); 90% of the time >1.5 fold (>5 copies per cell) >2 fold change (>1 copy per cell) R2 ≥ 0.95 7 X 105 Total Counts 6 positive and 8 negative in each reaction Genes Included ABL1 CCND3 CDK6 E2F4 MCM2 ORC5L RAD51 TGFB2 ANAPC1 CCNE1 CDK7 E2F5 MCM3 ORC6L RB1 TGFB3 CDKN1A E2F6 MCM4 PCNA RBBP8 TNXB ANAPC10 CCNE2 ANAPC11 CCNG2 CDKN1B EP300 MCM5 PIK3CA RBL1 TP53 ANAPC2 CCNH CDKN1C ESPL1 MCM6 PIK3R1 RBL2 TP73 ANAPC4 CDAN1 CDKN2A FBXW7 MCM7 PKMYT1 RBX1 TWIST1 ANAPC5 CDC14A CDKN2B FZR1 MDM2 PLK1 UBE2M RELA ANAPC7 CDC14B CDKN2C GADD45A MNAT1 POLA2 RPA1 WEE1 ARF1 CDC14C CDKN2D GADD45B MPEG1 POLE RPA2 XPO1 RPA3 YWHAB ARF3 CDC16 CFL1 GADD45G MPL POLE2 ATF6B CDC2 CHEK1 GBA2 MRE11A POLR1A RPS6KA1 YWHAE ATM CDC20 CHEK2 GSK3B MYC POLR1B SFN YWHAG ATR CDC23 CREB3 HDAC1 MYT1 POLR1C SHH YWHAH POLR1D SKP1 BRCA1 CDC25A CREB3L1 HDAC2 NACA BUB1 CDC25B CREB3L3 HDAC3 NACAP1 PRB1 SKP2 BUB1B CDC25C CREB3L4 HDAC4 NEDD8 PRIM1 SMAD2 BUB3 CDC27 CREBBP HDAC5 NEK1 PRIM2 SMAD3 CCNA1 CDC34 CUL1 HDAC6 NFKB1 PRKDC SMAD4 YWHAQ YWHAZ CCNA2 CDC45L DBF4 HDAC8 NFKBIA PTCH1 SMC1A CLTC* CCNB1 CDC6 DHFR JUN NXT1 PTPRA TBC1D8 GAPDH* CCNB2 CDC7 DTX4 MAD1L1 ORC1L PTTG1 TBX2 GUSB* CCNB3 CDH1 E2F1 MAD2L1 ORC2L PTTG2 TFDP1 HPRT1* CCND1 CDK2 E2F2 MAD2L2 ORC3L RAC1 TFDP2 PGK1* CCND2 CDK4 E2F3 MAPK8 ORC4L RAD50 TGFB1 TUBB* phases of the cell cycle. The verification was done using MSigDB, a repository of gene expression data developed by researchers at the Massachusetts Institute of Technology and the Broad Institute Inc (Subramanian, Tamayo, et al., 2005, PNAS 102, 15545-15550.). Other public databases were used to obtain functional gene expression information for each gene. The final nCounter Virtual Cell Cycle Gene Set consists of 183 cell cycle-related genes and six internal reference genes. For the gene list and additional information about this gene set, visit the nCounter Virtual Panels product page at www.nanostring.com. Nanostring acknowledges Ingenuity® Systems, Inc. (www.ingenuity.com) pathway tools used in the development of the gene list and supporting biological and chemical content. nCounter® Analysis System Overview The nCounter® Analysis System from NanoString offers a cost-effective way to easily profile hundreds of gene transcripts simultaneously with high sensitivity and precision. The digital detection of target molecules and high levels of multiplexing eliminate the compromise between data quality and data quantity, bringing better sensitivity, reproducibility, and linearity to your results. It is ideal for studying defined gene sets across a large sample set, e.g., microarray validation, pathway analysis, biomarker validation, and splice variation analysis. The system utilizes a novel digital technology that is based on direct multiplexed measurement of gene expression and offers high levels of precision and sensitivity (<1 copy per cell). The technology uses molecular “barcodes” and single molecule imaging to detect and count hundreds of unique transcripts in a single reaction. *Internal Reference Genes Ordering Information Description Quantity/Use P/N nCounter Virtual Cell Cycle Gene Set 48 Assays 96 Assays 192 Assays 384 Assays GXA-VCC1-048 GXA-VCC1-096 GXA-VCC1-192 GXA-VCC1-384 nCounter Master Kit All reagents, sample cartridges, and consumables necessary for processing 48 or 192 assays. 48 Assays 192 Assays NAA-AKIT-048 NAA-AKIT-192 NanoString Technologies, Inc. 530 Fairview Ave N, Suite 2000 Seattle, Washington 98109 (206) 378-NANO • (206) 378-6266 www.nanostring.com © 2009-2010 NanoString Technologies, Inc. All rights reserved. NanoString Technologies, NanoString, nCounter, and Molecules That Count are trademarks of NanoString Technologies, Inc., in the United States and/or other countries. All other trademarks and/or service marks not owned by NanoString that appear in this product data sheet are the property of their respective owners. v.20100629