* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download 幻灯片 1
Epigenetics in stem-cell differentiation wikipedia , lookup
Genome (book) wikipedia , lookup
Protein moonlighting wikipedia , lookup
Epigenetics of neurodegenerative diseases wikipedia , lookup
Designer baby wikipedia , lookup
Point mutation wikipedia , lookup
Cancer epigenetics wikipedia , lookup
Gene expression programming wikipedia , lookup
Epigenetics of diabetes Type 2 wikipedia , lookup
Primary transcript wikipedia , lookup
Oncogenomics wikipedia , lookup
Site-specific recombinase technology wikipedia , lookup
Epigenetics of human development wikipedia , lookup
Long non-coding RNA wikipedia , lookup
Vectors in gene therapy wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Nutriepigenomics wikipedia , lookup
Gene therapy of the human retina wikipedia , lookup
Gene expression profiling wikipedia , lookup
Polycomb Group Proteins and Cancer wikipedia , lookup
Therapeutic gene modulation wikipedia , lookup
Secreted frizzled-related protein 1 wikipedia , lookup
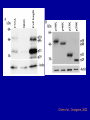
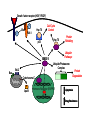
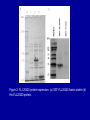
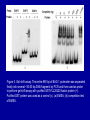
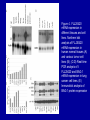
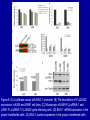
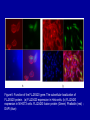
Cloning and functional analysis of FLJ20420: A novel BAG-1 promoter transcription factor 陈 军 天津市肺癌研究所 天津医科大学总医院肺部肿瘤外科 The structure of human BAG family proteins Li et al., Chin J Lung Cancer. 2009, BACKGROUND: • BAG-1 is a multifunctional protein that interacts with a wide range of target molecules to regulate apoptosis, proliferation, transcription, metastasis and mobility (Tang et al., 2002). • BAG-1 is widely over-expressed in various human malignancies and its expression may have clinical utility as a prognostic marker in early breast cancer (Tang et al., 2004). • However, most studies focus on the mechanism by which BAG-1 interacts with proteins to inhibit apoptosis. The molecules that regulate BAG-1 over-expression in malignant cells are currently unknown. Chen et al, Oncogene, 2002 Chen et al, Oncogene, 2002 Growth factor receptor (HGF, PDGF) P Ras P Hsp 70 Raf-1 Cell Cycle Control Hsp 70 p53 Ubiquitin Pathways BAG-1 Bax Protein Refolding Ubiquitin/Proteasome Complex Bcl-2 Cytochorome C Apaf-1 Hormone Receptor (ER/PR) Protein Degradation Apoptosis Drug Resistance Nucleotide sequence of human BAG-1 gene 5'- FLanking regions, putative transcription factor binding sites and CpG island Yang et al, Oncogene, 1998 Deletion analysis of the 5'-flanking region of the BAG-1 gene Yang et al, Oncogene, 1998 DESIGN: Our lab was the first to isolate the human BAG-1 promoter and characterize its functions (Yang et al., 1999). Since BAG-1 is highly over-expressed in human cervical cancer Hela cell line, we decided to identify transcription molecules regulating BAG-1 expression using BAG-1 promoter as a probe to screen Human Hela cDNA library λTripIEx by Southernwestern Blot Figure 1. Sequence analysis of positive clone cDNA and deduced amino acid sequence. Numbers indicate the positions of nucleotides. Figure 2: FLJ 20420 protein expression. (a) GST-FLJ20420 fusion protein (b) His-FLJ20420 protein. Figure 3. Gel shift assay. The entire 890 bp of BAG-1 protmoter was separated finally into several ~30-50 bp DNA fragment by PCR and then used as probe to perform gel shift assay with purified GST-FLJ20420 fusion protein (+). Purified GST protein was used as a control (v). (a) EMSA; (b) competition test of EMSA. Figure 4. FLJ20420 mRNA expression in different tissues and cell lines. Northern blot analysis of FLJ20420 mRNA expression in human normal tissues (A) and various tumor cell lines (B); (C-D) Real-time PCR analysis of t FLJ20420 and BAG-1 mRNA expression in lung cancer cell lines. (E) Immunoblot analysis of BAG-1 protein expression. Figure 5: (A) Luciferase assay with BAG-1 promoter. (B) The knockdown of FLJ20420 expression in A549 and L9981 cell lines. (C) Microarrays of A549-FLJ-siRNA-1 and L9981-FLJ-siRNA-1 FLJ20420 gene silencing cells. (D) BAG-1 mRNA expression in the proper transfected cells. (D) BAG-1 protein expression in the proper transfected cells . Figure 6: Function of the FLJ20420 gene. The subcellular localization of FLJ20420 protein. (a) FLJ20420 expression in Hela cells. (b) FLJ20420 expression in NIH/3T3 cells. FLJ20420 fusion protein (Green); Phalloidin (red) ; DAPI (blue) Figure 6: Function of the FLJ20420 gene. (B) Cell cycle analysis was evaluated by FACS after using PI to stain the cellular DNA. (C) Cell viability assay by the MTT assay. (D) Cells transfected with FLJ20420 siRNA were treated with cisplatin for 24 hours and then analyzed by the MTT assay. (E) Cell apoptosis assay by FACS. Gene expression profile of FLJ20420silenced A549 cells Microarray analysis revealed: 655 upregulated genes, including 272 known genes (272/38,500 = 0.706%). 619 downregulated genes, including 233 known genes (233/38,500 = 0.605%). Gene expression profile of FLJ20420-silenced A549 cells Pathway Gene Symbol Apoptosi AKT1, BCL2L1, CASP3, CASP7, CFLAR, CYCS, IL1RAP, PIK3CB, TNFRSF10A Calcium signaling pathway ADRB2, CAMK2D, ERBB2, MYLK, OXTR, PLCB4, PTK2B, VDAC3 cell cycle CCNE2, CDC2, CDK6, MAD1L1, TGFB2, ECM-receptor interaction COL5A1, ITGA11, ITGA2, SPP1, THBS1, VTN ErbB signaling pathway AKT1, CAMK2D, CBLB, ERBB2, GAB1, PIK3CB, RPS6KB1 Insulin signaling pathway AKT1, CBLB, MKNK2, PDE3A, PFKL, PIK3CB, PPP1R3C, PTPRF, RAPGEF1, RHEB, RHOQ, RPS6KB1 Jak-STAT signaling pathway AKT1, BCL2L1, CBLB, CSF2, IFNAR1, IL6, IL6R, IL6ST, JAK3, LEPR, PIAS1, PIK3CB MAPK signaling pathway AKT1, BDNF, CASP3, DUSP3, FGF2, FGFR2, JUND, MAP3K8, MKNK2, MRAS, PDGFA, PLA2G12A, PPM1A, RASGRP1, STK4, TAOK1, TGFB2, TGFBR1 p53 signaling pathway CASP3, CCNE2, CDC2, CDK6, CYCS, IGFBP3, PERP, PTEN, THBS1 TGF-beta signaling pathway ACVR2A, ID2, INHBC, RPS6KB1, SMAD5, TGFB2, TGFBR1, THBS1 Toll-like receptor signaling pathway AKT1, IFNAR1, IL6, IRF5, MAP3K8, PIK3CB, SPP1, TLR1, TRAF3 Wnt signaling pathway CAMK2D, PLCB4, PPP2R5E, TCF7L1, VANGL1, WNT6 Figure 7: Real-time PCR analysis of gene expression in A549 cells transfected with FLJ20420 siRNA. (A) The expression level changes of 15 genes shown to be upregulated (15/17). (B) The expression level changes of 12 genes shown to be downregulated (12/15) . Figure 8: FLJ20420 and BAG-1 expression in lung cancer tissues by Microarray (total 72 paired specimens, which included 29 adenocarcinomas and 43 squamous cell carcinomas). Summary • Identified and characterized a negative BAG-1 regulating transcription factor FLJ20420. • There is a high expression level of FLJ20420 in lung cancer tissues, compared to the paired lung tissues; • FLJ20420 plays an important role in the apoptosis and oncogenesis of lung caner. ACKNOWLEGEMENT • • • • • • • • Dr. Qinghua Zhou Dr. Hongyu Liu Dr. Zhihao Wu Dr. Ke Xu Dr. Haisu Wan Dr. Zhigang Li Wu Heng Staffs in Dept. of Lung Surgery • Staffs in Lung Cancer Institute Dr. Shou-Ching Tang Dr. Nian Zhang Dr. Allen Gao The grants: National Natural Science Foundation of China ; National “863” Plans National Sic-Tech Support Program Tianjin Sci-Tech Support Program Thank You!