* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download AllBio_DJK
Ridge (biology) wikipedia , lookup
Polycomb Group Proteins and Cancer wikipedia , lookup
Primary transcript wikipedia , lookup
Long non-coding RNA wikipedia , lookup
Transposable element wikipedia , lookup
Saethre–Chotzen syndrome wikipedia , lookup
Epigenomics wikipedia , lookup
Copy-number variation wikipedia , lookup
Metagenomics wikipedia , lookup
Minimal genome wikipedia , lookup
Gene nomenclature wikipedia , lookup
Frameshift mutation wikipedia , lookup
Pathogenomics wikipedia , lookup
Epigenetics of neurodegenerative diseases wikipedia , lookup
Cancer epigenetics wikipedia , lookup
Genomic imprinting wikipedia , lookup
Gene therapy wikipedia , lookup
No-SCAR (Scarless Cas9 Assisted Recombineering) Genome Editing wikipedia , lookup
Human genome wikipedia , lookup
Human genetic variation wikipedia , lookup
Epigenetics of diabetes Type 2 wikipedia , lookup
Genetic engineering wikipedia , lookup
Gene desert wikipedia , lookup
Non-coding DNA wikipedia , lookup
Vectors in gene therapy wikipedia , lookup
Oncogenomics wikipedia , lookup
Epigenetics of human development wikipedia , lookup
Public health genomics wikipedia , lookup
Gene expression programming wikipedia , lookup
Nutriepigenomics wikipedia , lookup
Genome (book) wikipedia , lookup
Genome editing wikipedia , lookup
Point mutation wikipedia , lookup
Helitron (biology) wikipedia , lookup
History of genetic engineering wikipedia , lookup
Gene expression profiling wikipedia , lookup
Genome evolution wikipedia , lookup
Therapeutic gene modulation wikipedia , lookup
Site-specific recombinase technology wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Microevolution wikipedia , lookup
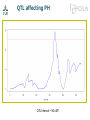
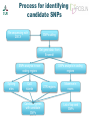
Wednesday from QTL to candidate genes Xidan Li Xiaodong Liu DJ de Koning Overview of today • Schedule for teaching day • Morning Lectures • 9:00 – 10:00 Lecture: Chasing the genetic basis of a QTL in chicken – DJ de Koning • 10:00 – 10:15 coffee break • 10:15 – 10:45 Lecture: Bioinformatics pipeline for targeted sequencing of QTL region – Xiaodong Liu • 10:45 – 11:00 leg stretcher • 11:00 – 11:30 Lecture: Identification and evaluation of causative genetic variants – Xidan Li • 11.30-12.00 discussion about morning topics • 12.00-13.00 lunch • • • • • Afternoon exercises 13:00 – 14:00 NGS data aligning 14:00 – 15:00 SNPs calling 15:00 – 16:00 Identify and evaluate causative genetic variants 16:00 – 17:00 evaluate results and questions Chasing the genetic basis of a QTL in chicken DJ de Koning [email protected] Contributors • Swedish University of Agricultural Sciences, Uppsala University • Xidan Li • Xiaodong Liu • Roslin Institute, University of Edinburgh • • • • Javad Nadaf Ian Dunn Chris Haley Ark-Genomics: Alison Downing, Mark Fell, Frances Turner • INRA, Unité de Recherches Avicoles • Cécile Berri • Elizabeth Le Bihan-Duval From sequence to consequence Phenotype Complex Traits The observed trait is sum of many genes and environmental factors Detection QTL using exotic crosses Use of of extreme crosses to unravel complex traits qq QQ Qq QQ Qq qq Detection of ”QTL” • Use heritable variation in the genome as DNA Markers • Follow inheritance of DNA markers through population • Compare inheritance pattern with character of interest Quantitative trait locus (QTL) • Region of the genome with a ’significant’ effect on our trait of interest. • Large region with very many genes. Intermezzo: tool for QTL analysis www.gridqtl.org.uk Nowadays: Association studies • Take a large (thousands), representative, sample of the population • Characterise for a very large number of DNA variants • Estimate a putative effect of every DNA variant on the trait of interest Challenge remains: What is the gene? • Very large area • Very noisy signal • Many candidate genes • Signal may not mark the gene Livestock genomics Output •QTL: Animal QTLdb http://www.animalgenome.org/QTLdb/ • Chicken 3162 QTL from 158 papers • Pigs • From QTL to QTN • Pigs IGF2 • Cattle DGAT1, ABCG2 6818 QTL from 290 papers … • Cattle 5920 QTL from 330 papers 1000’s of QTL, very few QTN Next step up: Gene expression studies • Measure the expression of thousands of genes simultaneously • Snapshot of what is happening in a given tissue at a given time. X 50 Test Statistic 40 30 20 10 0 0 50 100 150 cM • QTL study AND gene expression study in Population. • What are the gene expression effects of this QTL 50 Test Statistic 40 30 20 10 0 0 50 100 150 cM eQTL: Genome region that affects gene expression Targeted eQTL Mapping • Focus expression analysis on most informative individuals • eQTL underlying functional QTL • Increased power for target regions Application to a chicken QTL PH in chicken meat • Very important meat quality trait • Related to activity on the slaughter line • Here measured 15 minutes post mortem Chicken High growth Line, Low growth Line F. Ricard, 1975 Nadaf et al 2007 QTL affecting PH QTL Interval ~ 50 cM? Experimental design • What are the local and global effects of this QTL on gene expression? • Identify 12 birds with QQ genotypes on the basis of flanking markers and 12 with qq genotype • Perform microarray analyis using mRNA from breast muscle (P. Major) • Agilent 44k Array: 2-colour, dye-balanced Targeted genetical genomic approach 700 F2 24 F2 12 QQ 12 qq 12 Microarray chips (Agilent 44k) RNA RNA Enriched signals at the QTL position Closer look at the QTL area QTL appears to act on a region < 1Mb Top 10 t P.Value adj.P.Val Alternative Gene name CR385747 RCJMB04_23c A_87_P014348 19 -16.51 2.87E-10 7.19E-06 ZFY -16.28 3.42E-10 7.19E-06 ACOT9 A_87_P030344 BU299642 -12.64 8.27E-09 0.0001 PRDX4 A_87_P034725 BU106729 11.03 4.38E-08 0.0005 A_87_P014256 CR390282 10.28 1.02E-07 0.0009 KLHL15 A_87_P011383 CR523763 9.25 3.57E-07 0.0025 KLHL15 A_87_P032384 BU230994 8.44 1.03E-06 0.0062 PRDX4 A_87_P006189 TC202659 7.22 5.78E-06 0.0304 MSL3L1 A_87_P025536 BU476093 7.04 7.56E-06 0.0353 APOO A_87_P034683 BU108463 6.89 9.55E-06 0.0401 PRDX4 ProbeName GeneName A_87_P016951 Enriched signals at the QTL position • 16 differentially expressed probes in 1Mb region around QTL • QTL acting at chromatin or methylation level? • PH simply one of the downstream effects. Next Step: Re-Sequencing the QTL region • 5 birds of each QTL genotype • Selected DNA from 1 Mb around QTL with Agilent SureSelect Target Enrichment • One lane on Illumina GA flow cell: 151 bp paired-end • 4.9 Gbase of raw sequencing reads • ~200 x coverage of each individual chicken To be continued • YOU will work with this NGS data today! • The work up to the NGS has been published Over to you! • Then coffee Process for identifying candidate SNPs Re-sequencing with 200 X SNPs calling Get gene data from Ensembl SNPs analysis in noncoding regions Splicing sites CpG islands Candidate genes with candidate SNPs SNPs analysis in coding regions UTR regions Missense in exons List of top rank SNPs Non-synonymous mutations ACOT9 Description: acyl-Coenzyme A thioesterase 2, mitochondrial [Source:RefSeq peptide;Acc:NP_001012841] Mutations: QQ coordinate in chr 121615732 position in proteins 290 ref codon VS. Mutated codon [ TAC / TAT V / I ] 121615675 121615732 309 290 [ CGC / CGA A / S ] [ TAC / TAT V / I ] qq Mutations predictions: A - S (309) V - I (290) conservaton score 0.08 0.09 physico-chemical properties 0.54 0.201 final effect 0.3726 0.201 All scores are in range (0 - 1). The higher score, the more effect to protein function. ACOT9 • Most significant gene from eQTL study • Mitochondrial gene • Function of this particular gene not clear. • “Acyl-CoA thioesterases are a group of enzymes that catalyze the hydrolysis of acyl-CoAs to the free fatty acid and coenzyme A (CoASH), providing the potential to regulate intracellular levels of acyl-CoAs, free fatty acids and CoASH.” F1NR19 Description: DNA polymerase Source: UniProtKB/TrEMBL F1NR19 Mutations: QQ coordinate in chr 121338134 121464309 121466769 121478463 position in proteins 1396 523 466 289 ref codon VS. Mutated codon [ CGT / CAT T / M ] [ AGG / AAG P / L ] [ CGC / CAC A / V ] [ ATG / ACG H / R ] 121464309 121466769 523 466 [ AGG / AAG P / L ] [ CGC / CAC A / V ] qq Mutations predictions: H - R (289) A - V (466) P - L (523) T-M (1396) conservaton score 0.04 0.05 0.11 physico-chemical properties 0.543 0.304 0.703 final effect 0.02172 0.0152 0.07733 0.03 0.412 0.01236 ENSGALT0000026337 PRDX4 • Peroxiredoxin 4 • Antioxidant enzyme, regulates NFĸB • Highly differentially expressed but no candidate SNPs • 2 probes up, 1 down => Splicing? • Still a strong functional candidate