* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download week2
Epigenetics of human development wikipedia , lookup
Genomic imprinting wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Genetic drift wikipedia , lookup
Koinophilia wikipedia , lookup
Genome evolution wikipedia , lookup
Pharmacogenomics wikipedia , lookup
Dual inheritance theory wikipedia , lookup
Medical genetics wikipedia , lookup
Site-specific recombinase technology wikipedia , lookup
Polymorphism (biology) wikipedia , lookup
Gene expression profiling wikipedia , lookup
Gene expression programming wikipedia , lookup
Genetic testing wikipedia , lookup
Genetic engineering wikipedia , lookup
History of genetic engineering wikipedia , lookup
Biology and consumer behaviour wikipedia , lookup
Human genetic variation wikipedia , lookup
Behavioural genetics wikipedia , lookup
Population genetics wikipedia , lookup
Public health genomics wikipedia , lookup
Designer baby wikipedia , lookup
Heritability of IQ wikipedia , lookup
Genome (book) wikipedia , lookup
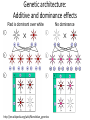
What is a QTL? • Quantitative trait locus (loci) • Region of chromosome that contributes to variation in a quantitative trait • Generally used to study “complex traits”, i.e., controlled by many genes and environmental factors Why would you want to map QTL? • Identify genes responsible for variation, e.g., • Medicine – disease susceptibility, reaction to drugs • Agriculture – crop/livestock improvement • Evolution Why would you want to map QTL? • Identify genes responsible for variation • Understand genetic architecture What is genetic architecture? • Number of loci that contribute to a trait • Distribution of effect sizes • “Mode of action” of loci Genetic architecture: Number of loci • Number of loci contributing to differences in a trait between two lines/ strains • Historically, estimated in various ways, especially the Castle-Wright index/ estimator • Castle-Wright index assumes – Two homozygous parents are crossed, one only has increasing alleles and the other only has decreasing alleles for the trait – All loci affect the trait equally – Loci affecting the trait are unlinked – No dominance or epistasis • More modern methods avoid some of these assumptions Genetic architecture: Distribution of effect sizes Behavioural traits Non-behavioural traits Flint and Mott 2008; Nature 456: 724 Genetic architecture: Additive and dominance effects Red is dominant over white http://en.wikipedia.org/wiki/Mendelian_genetics No dominance Why care about genetic architecture? Can the identification of QTL useful if you do not identify the underlying genes? • How big are the largest effect sizes? Is a QTL worth pursuing? • Why are traits correlated? Do they share QTL (pleiotropy)? E.g., • Medicine – QTL for reading disability and ADHD Why care about genetic architecture? • Evolution – adaptation, e.g., Peichel et al 2001, Nature 414: 901-905 Why care about genetic architecture? • Evolution – speciation, e.g., Hawthorne and Via 2001, Nature 412: 905-907 Why care about genetic architecture? • Evolution – do QTL from different studies colocalize? Marker assisted selection (MAS) in agriculture • Advantages/disadvantages QTL mapping vs. other strategies • What is the question? – Which genes contribute to variation? – Which genes contribute to trait? QTL mapping vs. other strategies • • • • QTL mapping Candidate gene studies Mutagenesis Microarray, serial analysis of gene expression (SAGE) – gene vs. network focus (Flint and Mott 2008, Nature 456: 724-727) • Other?