* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download Amidase overexpression - Duke Trinity College of Arts and Sciences
Survey
Document related concepts
Biochemical switches in the cell cycle wikipedia , lookup
Cell encapsulation wikipedia , lookup
Cell membrane wikipedia , lookup
Signal transduction wikipedia , lookup
Extracellular matrix wikipedia , lookup
Endomembrane system wikipedia , lookup
Cell culture wikipedia , lookup
Cellular differentiation wikipedia , lookup
Organ-on-a-chip wikipedia , lookup
Programmed cell death wikipedia , lookup
Cell growth wikipedia , lookup
Transcript
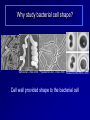
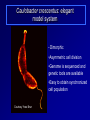
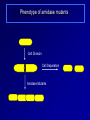
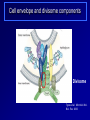
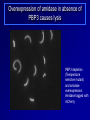
Antibiotics Resistance: Lessons learned from Bacterial Cell Division Richa Priyadarshini Assistant Professor Dept. of Life Sciences School of Natural Sciences Shiv Nadar University Urbanization? On September 18 this year the Obama Administration announced the White House National Strategy for Combating Antibiotic Resistant Bacteria (CARB). The strategy outlines bold steps to slow the public health threat of antibiotic resistant bacteria, including efforts to stimulate innovative research. Study of Antibiotic Resistance Two Approaches towards studying antibiotic resistance: 1. Study of bacterial cell wall biogenesis using model organism (E. coli and Caulobacter) 2. Use on metagenomics It's not in the open we feel comforted but in the shadows. … We can't feel at home with the infinite sky above and around us. Space must be cut off, shaped, defined, for us to inhabit. From cradle to coffin, it's enclosure that defines us. —Robert Morgan Why study bacterial cell shape? Varma et al. J. Bac. 2004 Priyadarshini et al. J. Bac. 2006 Kruse et al. Mol. Micro. 2004 Cell wall provided shape to the bacterial cell Bacterial Cell Wall Targets of cell wall-active antibiotics. McCallum N et al. Antimicrob. Agents Chemother. 2011;55:1391-1402 Mechanism Leading to Antibiotic Resistance Genetic mutations lead to: changes in binding proteins ribosomes membrane structure inactivating enzymes Conly J CMAJ 2002;167:885-891 A population-based antibiotic-resistance mechanism in bacteria. HH Lee et al. Nature 467, 82-85 (2010) doi:10.1038/nature09354 Strategies for control of antibiotic resistance Minimize the use of antibiotic improving infection control developing new antibiotics Global policy for tracking the emergence of antibiotic resistance Cell wall modifying enzymes (Penicillin Binding Proteins) 1 PBP 2/ PBP 3 PBP 1A/ 1B/1C Holtje et al. MMBR Mar 1998 Cell wall modifying enzymes (Hydrolases) D,D-carboxypeptidae PBP5 & PBP6 1 Amidase Endopeptidase PBP4 &PBP7 Holtje et al. MMBR Mar 1998 Hydrolases as antibiotic targets MMBR Mar 1998 Hydrolases as antibiotic targets Penicillin (Beta-Lactams) Vancomycin MMBR Mar 1998 How is the activity of cell wall hydrolases regulated? Penicillin Vancomycin Bacitracin MMBR Mar 1998 How is the activity of cell wall hydrolases regulated? Penicillin Vancomycin Bacitracin fosfomycin Cycloserine MMBR Mar 1998 Caulobacter crescentus: elegant model system • Dimorphic •Asymmetric cell division •Genome is sequenced and genetic tools are available •Easy to obtain synchronized cell population Courtesy Yves Brun Cell wall modifying enzymes (Hydrolases) 1 Amidase E. coli has 3 amidases: AmiA, AmiB and AmiC Phenotype of amidase mutants Cell Division Cell Separation Amidase Mutants What is the role of amidase in Caulobacter cells? Localization of amidase in Caulobacter Ami-mCherry Caulobacter has only one annotated amidase Cell envelope and divisome components Divisome Typas et al. Microbiol. Mol. Biol. Rev. 2006 Amidase localizes to mid-cell after FtsN 0min FtsN-YFP Ami-mCherry Merge 15min 30min 45min 60min 75min 90min What happens when amidase activity is removed from the cell Xylose inducible promoter system No Xylose Off Xylose ON -Xylose promoter -Gene of interest Amidase depletion causes cell division defects in Caulobacter Effect of over-activity of amidase on Caulobacter cells Overexpression of amidase causes cell chaining and filamentation Time after induction 6hr 7hr 8hr 10hr 0.2% glu 0.3% xyl Amidase overexpression Overexpression of amidase causes cell chaining Amidase overexpression Time lapse at 30°C with xylose Balance between cell wall degradation and synthesis is disturbed in amidase overexpression strains Wild Type Amidase overexpression PBP3 Amidase Cell wall Overexpression of amidase in absence of PBP3 causes lysis PBP3 depletion (Temperature sensitive mutant) and amidase overexpression. Amidase tagged with mCherry Summary Amidase is essential for viability in Caulobacter Amidase depletion and overexpression causes cell division defects and lysis of cells Amidase overexpression disturbs the delicate balance between synthesis and hydrolysis of the septal peptidoglycan Role of environment in antibiotic resistance Ecology of Antibiotics Bacteria found in soil contain antibiotic resistance genes D'Costa et al., Science. 311 (5759): 374-377 Spread of antibiotic resistance Antibiotics are communication molecules of bacteria Humans communicate with poetry, bacteria…. www.sciencedaily.com Metagenomic Approaches to Combat antibiotic resistance metagenomic tools to understand the microbial composition of certain unique environments identify the presence of antibiotic resistance genes and pathways identify pathways and genes responsible for production of novel antimicrobial compounds Urbanization? Acknowledgements Lab members: Ph.D. Students- Deepika Chauhan and Amrita Dubey Vinita Tomar (Lab Assistant) Dept. Life Sciences Dr. Rupamanjari Ghosh (Director SONS) Shiv Nadar University Duke University Collaborators Christine Jacobs-Wagner (Yale University) School of Natural Sciences Environmental and Natural Resources Management Jyoti Sharma: http://snu.edu.in/naturalsciences/jyoti_kumar_sharma_profile.aspx Life Sciences Shailja Singh: http://snu.edu.in/naturalsciences/Shailja_Singh_profile.aspx Seema Sherawat : http://snu.edu.in/naturalsciences/Seema_Sehrawat_profile.aspx Anindita Chakrabarty: http://snu.edu.in/naturalsciences/anindita_chakrabarty.aspx Richa Priyadarshini: http://snu.edu.in/naturalsciences/Richa_Priyadarshini_profile.aspx School of Natural Sciences Dept. of Chemistry Subhabrata Sen: http://snu.edu.in/naturalsciences/Subhabrata_Sen_profile.aspx Gouriprasanna Roy: http://snu.edu.in/naturalsciences/gouriprasanna_roy_profile.aspx Bimlesh Lochab: http://snu.edu.in/naturalsciences/Bimlesh_Lochab_profile.aspx Parthapratim Munshi: http://snu.edu.in/naturalsciences/parthapratim_munshi_profile.aspx