* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download Exploring ClinVar
Survey
Document related concepts
Gene therapy wikipedia , lookup
Quantitative trait locus wikipedia , lookup
Pharmacogenomics wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Genome (book) wikipedia , lookup
Gene nomenclature wikipedia , lookup
Human genetic variation wikipedia , lookup
Neuronal ceroid lipofuscinosis wikipedia , lookup
Therapeutic gene modulation wikipedia , lookup
Dominance (genetics) wikipedia , lookup
BRCA mutation wikipedia , lookup
Microevolution wikipedia , lookup
Transcript
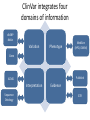
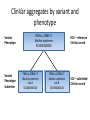
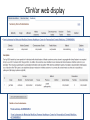
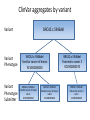
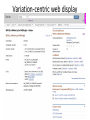
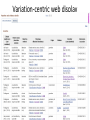
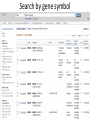
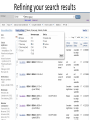
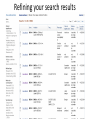
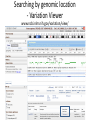
Exploring ClinVar: What's There and How Do I Use It? Melissa Landrum ICCG June 12, 2014 What is ClinVar? • Archive of assertions of clinical significance for variants relative to phenotypes • Fully public and freely available • Submission-driven database • Currently accepting variant/aggregate-level data; case-level support under development • Curation support from NCBI staff What is the difference between ClinVar and ClinGen? ClinGen ClinVar • An archival database at • The Clinical Genome Project – multiple NCBI institutions • Supported by NIH • Submit data to ClinVar intramural funding • Perform analysis on data extracted from ClinVar • Perform expert curation of variants and submit to ClinVar • Funded by grants from NHGRI ClinVar www.ncbi.nlm.nih.gov/clinvar/ ClinVar stats www.ncbi.nlm.nih.gov/clinvar/submitters/ ClinVar integrates four domains of information dbSNP dbVar Variation Phenotype MedGen (HPO, OMIM) Gene PubMed ACMG Interpretation Sequence Ontology Evidence GTR ClinVar aggregates by variant and phenotype Variant Phenotype Variant Phenotype Submitter FBN1:c.4786C>T Marfan syndrome RCV000000050 FBN1:c.4786C>T Marfan syndrome Lab A SCV000000010 FBN1:c.4786C>T Marfan syndrome Lab B SCV000000020 RCV – reference ClinVar record SCV – submitted ClinVar record ClinVar web display Clinical significance Benign Likely benign Uncertain significance Likely pathogenic Pathogenic Drug response Confers sensitivity Risk factor Association Protective Has conflicts ClinVar web display Review Status classified by single submitter classified by multiple submitters conflicting data from submitters not classified by submitter reviewed by expert panel reviewed by professional society ClinVar web display Accession • All records are accessioned and versioned • Supports search Allele summary • Track changes to the • Gene clinicaltype interpretation over • Variant time location • Genomic • • • • HGVS expressions* Molecular consequence* Links* Frequency* Phenotype summary • Names • Links* • Age of onset * • Prevalence * * May be provided by NCBI ClinVar web display ClinVar web display ClinVar aggregates by variant Variant Variant Phenotype Variant Phenotype Submitter BRCA2:c.5946del BRCA2:c.5946del Familiar cancer of breast RCV000000050 BRCA2:c.5946del Familial cancer of breast Lab A SCV000000010 BRCA2:c.5946del Familial cancer of breast Lab B SCV000000020 BRCA2:c.5946del Pancreatic cancer 2 RCV000000070 BRCA2:c.5946del Pancreatic cancer 2 Lab C SCV000000030 Variation-centric web display Variation-centric web display Variation-centric web display Searching ClinVar • • • • • Gene symbols HGVS expressions Protein changes Phenotypes Submitter Searching uses exact words and is not field-specific! Search by gene symbol Upcoming improvements to ClinVar searching • HGVS sensor – to restrict the query to variant name • Protein change sensor – to detect a partial protein change query, e.g. Arg911 • Phenotype query – to detect a phenotype term as query and provide the option to search for related terms Refining your search results Refining your search results Searching by genomic location - Variation Viewer www.ncbi.nlm.nih.gov/variation/view/ Accessing ClinVar data • Interactively on the web; updated weekly • Monthly full releases – Comprehensive XML extraction – VCF files – Tab-delimited summary files for genes, variants • E-utilities as web service or via command line • Annotation on graphic sequence displays • Variation Viewer www.ncbi.nlm.nih.gov/variation/view/ • Variation Reporter www.ncbi.nlm.nih.gov/variation/tools/reporter Acknowledgements Alex Astashyn Garth Brown Chao Chen Shanmuga Chitipiralla Arkadi Doubintchik Brad Holmes John Garner Anna Glodek Baoshan Gu Tim Hefferon Douglas Hoffman Wonhee Jang Brandi Kattman Ken Katz Jennifer Lee Donna Maglott Adriana Malheiro Michael Ovetsky Lon Phan George Riley Wendy Rubinstein Amanjeev Sethi Ray Tully Ricardo Villamarin Ming Ward Jim Ostell Steve Sherry Contact us at [email protected]