* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download Lecture #3 Date
Survey
Document related concepts
Tissue engineering wikipedia , lookup
Microtubule wikipedia , lookup
Cell growth wikipedia , lookup
Cellular differentiation wikipedia , lookup
Cell encapsulation wikipedia , lookup
Cell culture wikipedia , lookup
Cell nucleus wikipedia , lookup
Signal transduction wikipedia , lookup
Cytoplasmic streaming wikipedia , lookup
Organ-on-a-chip wikipedia , lookup
Extracellular matrix wikipedia , lookup
Cell membrane wikipedia , lookup
Cytokinesis wikipedia , lookup
Transcript
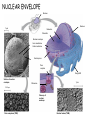
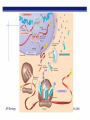
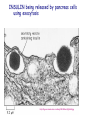
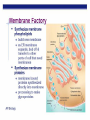
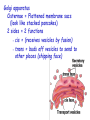
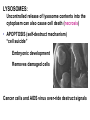
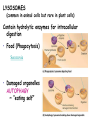
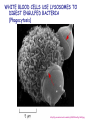
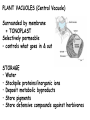
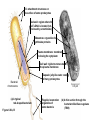
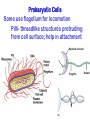
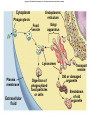
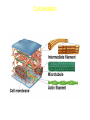
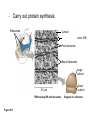
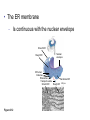
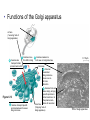
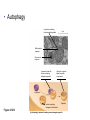
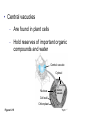
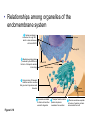
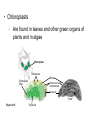
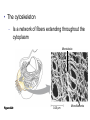
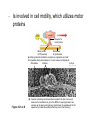
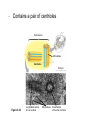
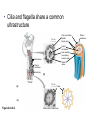
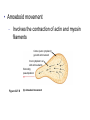
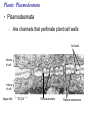
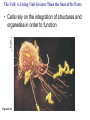
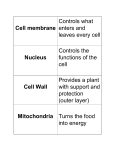
AP BIOLOGY Chapter 6 Cell Structure & Function WHAT’S NEW you didn’t learn in BIO I? Slide shows combined and modified from: http://gbs.glenbrook.k12.il.us/Academics/gbssci/bio/apbio/Lecture/lecture.htm; http://www.explorebiology.com/ http://home.att.net/~tljackson/neville.html NUCLEAR ENVELOPE http://cellbio.utmb.edu/cellbio/nuclear_envelope.htm DOUBLE MEMBRANE is fused in spots forming NUCLEAR PORES NUCLEAR LAMINA- netlike array of protein filaments on nuclear side of envelope that maintains the shape of the nucleus (Play a role in reforming nuclear membrane after cell division, if you inject antibodies to lamina proteins, nucleus can’t reform after mitosis) NUCLEAR ENVELOPE Nucleus Nucleus 1 µm Nucleolus Chromatin Nuclear envelope: Inner membrane Outer membrane Nuclear pore Pore complex Rough ER Surface of nuclear envelope. 1 µm Ribosome 0.25 µm Close-up of nuclear envelope Figure 6.10 Pore complexes (TEM). Nuclear lamina (TEM). ENDOMEMBRANE SYSTEM Regulates protein traffic and performs metabolic functions in the cell Includes: Plasma membrane Nuclear membrane Endoplasmic reticulum Golgi apparatus Vacuoles Lysosomes INSULIN being released by pancreas cells using exocytosis http://fig.cox.miami.edu/~cmallery/255/255ion/fig14x26.jpg Golgi apparatus Cisternae = Flattened membrane sacs (look like stacked pancakes) 2 sides = 2 functions cis = (receives vesicles by fusion) trans = buds off vesicles to send to other places (shipping face) Animation from: http://www.franklincollege.edu/bioweb/A&Pfiles/week04.html See a Golgi movie EVERYTHING’S CONNNECTED! LYSOSOMES: Uncontrolled release of lysosome contents into the cytoplasm can also cause cell death (necrosis) • APOPTOSIS (self-destruct mechanism) “cell suicide” Embryonic development Removes damaged cells Cancer cells and AIDS virus over-ride destruct signals LYSOSOMES (common in animal cells but rare in plant cells) Contain hydrolytic enzymes for intracellular digestion • Food (Phagocytosis) See movie • Damaged organelles AUTOPHAGY ~ “eating self” WHITE BLOOD CELLS USE LYSOSOMES TO DIGEST ENGULFED BACTERIA (Phagocytosis) http://fig.cox.miami.edu/~cmallery/255/255ion/fig14x28.jpg PEROXISOMES Other digestive enzyme sacs in both plants and animals In fat storing seeds (called GLYOXYSOMES) Break down fatty acids → sugars transport to mitochondria for energy In LIVER CELLS Detoxify alcohol & other poisons PRODUCE HYDROGEN PEROXIDE (TOXIN) but have enzyme (CATALASE) to break this down H2O2 → H2O + O2 PLASTIDS CHLOROPLASTS- contain pigment chlorophyll for photosynthesis CHROMOPLASTS- contain pigments that give fruits and flowers colors AMYLOPLASTS- store starch (amylose) in roots and tubers (colorless) http://www.jonathanwald.com/800x600/images/Red-Apple.jpg http://en.wikipedia.org/wiki/Image:Potato_-_Amyloplasts.jpg ANIMAL VACUOLES & VESICLES “transport vehicles” • FOOD VACUOLES Phagocytosis fuse with lysosomes • CONTRACTILE VACUOLES Freshwater organisms pump out excess water http://www.microscopy-uk.org.uk/mag/imgjun99/vidjun1.gif PLANT VACUOLES (Central Vacuole) Surrounded by membrane = TONOPLAST Selectively permeable – controls what goes in & out STORAGE • Water • Stockpile proteins/inorganic ions • Deposit metabolic byproducts • Store pigments • Store defensive compounds against herbivores 25 26 27 Pili: attachment structures on the surface of some prokaryotes Nucleoid: region where the cell’s DNA is located (not enclosed by a membrane) Ribosomes: organelles that synthesize proteins Plasma membrane: membrane enclosing the cytoplasm Cell wall: rigid structure outside the plasma membrane Capsule: jelly-like outer coating of many prokaryotes Bacterial chromosome (a) A typical rod-shaped bacterium Figure 6.6 A, B 0.5 µm Flagella: locomotion organelles of some bacteria (b) A thin section through the bacterium Bacillus coagulans (TEM) Prokaryotic Cells Some use flagellum for locomotion – Pilli- threadlike structures protruding from cell surface; help in attachment Bacterial cell wall Rotary motor Flagellin Sheath Copyright © The McGraw-Hill Companies, Inc. Permission required for reproduction or display. Cytoplasm Endoplasmic reticulum Phagocytosis Food vesicle Golgi apparatus Lysosomes Plasma membrane Extracellular fluid Digestion of phagocytized food particles or cells Transport vesicle Old or damaged organelle Breakdown of old organelle Chloroplasts •Chloroplasts are larger and more complex than mitochondria •Grana – closed compartments of stacked membranes •Thylakoids – disc shaped structure – light capturing pigment •Stroma – fluid matrix Cytoskeleton • Network of protein fibers supporting cell shape and anchoring organelles – Actin filaments – Microtubules – cell movement Microtubules Intermediate filaments Hollow tubes Facilitate cell movement Centrioles – barrel shaped organelles occur in pairs – help assemble animal cell’s microtubules Intermediate filaments Stable - don’t break down Actin Cytoskeleton Plant Cells • • Central vacuole – often found in the center of a plant, and serves as a storage facility for water and other materials Cell wall – primary walls – laid down while cell is growing – middle lamella – glues cells together – secondary walls – inside the primary cell walls after growth Plant Cell Animal Cells • Animal cells lack cell walls. – form extracellular matrix provides support, strength, and resilience Modified from: http://www.coe.unt.edu/ubms/documents/classnotes/ Fall2005/Chapter%205%20-%20Cell%20Structure.ppt Endomembrane system, II Golgi apparatus •ER products are modified, stored, and then shipped Cisternae: flattened membranous sacs trans face (shipping) & cis face (receiving) Transport vesicles Endomembrane system, III Lysosomes •sac of hydrolytic enzymes; digestion of macromolecules Phagocytosis Autophagy: recycle cell’s own organic material Tay-Sachs disease~ lipid-digestion disorder Endomembrane system, IV Vacuoles •membrane-bound sacs (larger than vesicles) Food (phagocytosis) Contractile (pump excess water) Central (storage in plants) •tonoplast membrane • Eukaryotic cells – Contain a true nucleus, bounded by a membranous nuclear envelope – Are generally quite a bit bigger than prokaryotic cells • A smaller cell – Has a higher surface to volume ratio, which facilitates the exchange of materials into and out of the cell Surface area increases while total volume remains constant 5 1 1 Figure 6.7 Total surface area (height width number of sides number of boxes) 6 150 750 Total volume (height width length number of boxes) 1 125 125 Surface-to-volume ratio (surface area volume) 6 12 6 A Panoramic View of the Eukaryotic Cell • Eukaryotic cells – Have extensive and elaborately arranged internal membranes, which form organelles • Plant and animal cells – Have most of the same organelles EVERYTHING’S CONNNECTED! The Nucleus: Genetic Library of the Cell • The nucleus – Contains most of the genes in the eukaryotic cell Ribosomes: Protein Factories in the Cell • Ribosomes – Are particles made of ribosomal RNA and protein – Carry out protein synthesis Ribosomes ER Cytosol Endoplasmic reticulum (ER) Free ribosomes Bound ribosomes Large subunit 0.5 µm TEM showing ER and ribosomes Figure 6.11 Small subunit Diagram of a ribosome Concept 6.4: The endomembrane system regulates protein traffic and performs metabolic functions in the cell • The endomembrane system – Includes many different structures The Endoplasmic Reticulum: Biosynthetic Factory • The endoplasmic reticulum (ER) – Accounts for more than half the total membrane in many eukaryotic cells • The ER membrane – Is continuous with the nuclear envelope Smooth ER Rough ER Nuclear envelope ER lumen Cisternae Ribosomes Transitional ER Transport vesicle Smooth ER Rough ER 200 µm Figure 6.12 • There are two distinct regions of ER – Smooth ER, which lacks ribosomes – Rough ER, which contains ribosomes Functions of Smooth ER • The smooth ER – Synthesizes lipids – Metabolizes carbohydrates – Stores calcium – Detoxifies poison Functions of Rough ER • The rough ER – Has bound ribosomes – Produces proteins and membranes, which are distributed by transport vesicles The Golgi Apparatus: Shipping and Receiving Center • The Golgi apparatus – Receives many of the transport vesicles produced in the rough ER – Consists of flattened membranous sacs called cisternae • Functions of the Golgi apparatus include – Modification of the products of the rough ER – Manufacture of certain macromolecules • Functions of the Golgi apparatus Golgi apparatus cis face (“receiving” side of Golgi apparatus) 1 Vesicles move 2 Vesicles coalesce to 6 Vesicles also form new cis Golgi cisternae from ER to Golgi transport certain Cisternae proteins back to ER 3 Cisternal maturation: Golgi cisternae move in a cisto-trans direction Figure 6.13 5 Vesicles transport specific proteins backward to newer Golgi cisternae 0.1 0 µm 4 Vesicles form and leave Golgi, carrying specific proteins to other locations or to the plasma membrane for secretion trans face (“shipping” side of Golgi apparatus) TEM of Golgi apparatus Lysosomes: Digestive Compartments • A lysosome – Is a membranous sac of hydrolytic enzymes – Can digest all kinds of macromolecules • Lysosomes carry out intracellular digestion by – Phagocytosis Nucleus 1 µm Lysosome Lysosome contains active hydrolytic enzymes Food vacuole fuses with lysosome Hydrolytic enzymes digest food particles Digestive enzymes Lysosome Plasma membrane Digestion Food vacuole Figure 6.14 A (a) Phagocytosis: lysosome digesting food • Autophagy Lysosome containing two damaged organelles 1µm Mitochondrion fragment Peroxisome fragment Lysosome fuses with vesicle containing damaged organelle Hydrolytic enzymes digest organelle components Lysosome Vesicle containing damaged mitochondrion Figure 6.14 B Digestion (b) Autophagy: lysosome breaking down damaged organelle Vacuoles: Diverse Maintenance Compartments • A plant or fungal cell – May have one or several vacuoles • Food vacuoles – Are formed by phagocytosis • Contractile vacuoles – Pump excess water out of protist cells • Central vacuoles – Are found in plant cells – Hold reserves of important organic compounds and water Central vacuole Cytosol Tonoplast Nucleus Central vacuole Cell wall Chloroplast Figure 6.15 5 µm The Endomembrane System: A Review • The endomembrane system – Is a complex and dynamic player in the cell’s compartmental organization • Relationships among organelles of the endomembrane system 1 Nuclear envelope is connected to rough ER, which is also continuous with smooth ER Nucleus Rough ER 2 Membranes and proteins produced by the ER flow in the form of transport vesicles to the Golgi Smooth ER cis Golgi Nuclear envelop 3 Golgi pinches off transport Vesicles and other vesicles that give rise to lysosomes and Vacuoles Plasma membrane trans Golgi 4 Lysosome available for fusion with another vesicle for digestion Figure 6.16 5 Transport vesicle carries 6 proteins to plasma membrane for secretion Plasma membrane expands by fusion of vesicles; proteins are secreted from cell • Concept 6.5: Mitochondria and chloroplasts change energy from one form to another • Mitochondria – Are the sites of cellular respiration • Chloroplasts – Found only in plants, are the sites of photosynthesis Mitochondria: Chemical Energy Conversion • Mitochondria – Are found in nearly all eukaryotic cells • Mitochondria are enclosed by two membranes – A smooth outer membrane – An inner membrane folded into cristae Mitochondrion Intermembrane space Outer membrane Free ribosomes in the mitochondrial matrix Inner membrane Cristae Matrix Figure 6.17 Mitochondrial DNA 100 µm Chloroplasts: Capture of Light Energy • The chloroplast – Is a specialized member of a family of closely related plant organelles called plastids – Contains chlorophyll • Chloroplasts – Are found in leaves and other green organs of plants and in algae Chloroplast Ribosomes Stroma Chloroplast DNA Inner and outer membranes Granum 1 µm Figure 6.18 Thylakoid • Chloroplast structure includes – Thylakoids, membranous sacs – Stroma, the internal fluid Peroxisomes: Oxidation • Peroxisomes – Produce hydrogen peroxide and convert it to water Chloroplast Peroxisome Mitochondrion Figure 6.19 1 µm • The cytoskeleton – Is a network of fibers extending throughout the cytoplasm Microtubule Figure 6.20 0.25 µm Microfilaments Roles of the Cytoskeleton: Support, Motility, and Regulation • The cytoskeleton – Gives mechanical support to the cell – Is involved in cell motility, which utilizes motor proteins ATP Vesicle Receptor for motor protein Motor protein Microtubule (ATP powered) of cytoskeleton (a) Motor proteins that attach to receptors on organelles can “walk” the organelles along microtubules or, in some cases, microfilaments. Vesicles Microtubule 0.25 µm Figure 6.21 A, B (b) Vesicles containing neurotransmitters migrate to the tips of nerve cell axons via the mechanism in (a). In this SEM of a squid giant axon, two vesicles can be seen moving along a microtubule. (A separate part of the experiment provided the evidence that they were in fact moving.) Components of the Cytoskeleton • There are three main types of fibers that make up the cytoskeleton Table 6.1 Microtubules • Microtubules – Shape the cell – Guide movement of organelles – Help separate the chromosome copies in dividing cells Centrosomes and Centrioles • The centrosome – Is considered to be a “microtubule-organizing center” – Contains a pair of centrioles Centrosome Microtubule Centrioles 0.25 µm Figure 6.22 Longitudinal section of one centriole Microtubules Cross section of the other centriole Cilia and Flagella • Cilia and flagella – Contain specialized arrangements of microtubules – Are locomotor appendages of some cells • Flagella beating pattern (a) Motion of flagella. A flagellum usually undulates, its snakelike motion driving a cell in the same direction as the axis of the flagellum. Propulsion of a human sperm cell is an example of flagellatelocomotion (LM). Direction of swimming Figure 6.23 A 1 µm • Ciliary motion (b) Motion of cilia. Cilia have a backand-forth motion that moves the cell in a direction perpendicular to the axis of the cilium. A dense nap of cilia, beating at a rate of about 40 to 60 strokes a second, covers this Colpidium, a freshwater protozoan (SEM). Figure 6.23 B 15 µm • Cilia and flagella share a common ultrastructure Outer microtubule doublet Dynein arms 0.1 µm Central microtubule Outer doublets cross-linking proteins inside Microtubules Radial spoke Plasma membrane Basal body (b) 0.5 µm (a) 0.1 µm Triplet (c) Figure 6.24 A-C Cross section of basal body Plasma membrane • The protein dynein – Is responsible for the bending movement of cilia and flagella Microtubule doublets ATP Dynein arm (a) Powered by ATP, the dynein arms of one microtubule doublet grip the adjacent doublet, push it up, release, and then grip again. If the two microtubule doublets were not attached, they would slide relative to each other. Figure 6.25 A ATP Outer doublets cross-linking proteins Anchorage in cell (b) In a cilium or flagellum, two adjacent doublets cannot slide far because they are physically restrained by proteins, so they bend. (Only two of the nine outer doublets in Figure 6.24b are shown here.) Figure 6.25 B 1 3 2 (c) Localized, synchronized activation of many dynein arms probably causes a bend to begin at the base of the Cilium or flagellum and move outward toward the tip. Many successive bends, such as the ones shown here to the left and right, result in a wavelike motion. In this diagram, the two central microtubules and the cross-linking proteins are not shown. Figure 6.25 C Microfilaments (Actin Filaments) • Microfilaments – Are built from molecules of the protein actin – Are found in microvilli Microvillus Plasma membrane Microfilaments (actin filaments) Intermediate filaments Figure 6.26 0.25 µm • Microfilaments that function in cellular motility – Contain the protein myosin in addition to actin Muscle cell Actin filament Myosin filament Myosin arm Figure 6.27 A (a) Myosin motors in muscle cell contraction. • Amoeboid movement – Involves the contraction of actin and myosin filaments Cortex (outer cytoplasm): gel with actin network Inner cytoplasm: sol with actin subunits Extending pseudopodium Figure 6.27 B (b) Amoeboid movement • Cytoplasmic streaming – Is another form of locomotion created by microfilaments Nonmoving cytoplasm (gel) Chloroplast Streaming cytoplasm (sol) Parallel actin filaments Figure 6.27 C (b) Cytoplasmic streaming in plant cells Cell wall Intermediate Filaments • Intermediate filaments – Support cell shape – Fix organelles in place Concept 6.7: Extracellular components and connections between cells help coordinate cellular activities Cell Walls of Plants • The cell wall – Is an extracellular structure of plant cells that distinguishes them from animal cells • Plant cell walls – Are made of cellulose fibers embedded in other polysaccharides and protein – May have multiple layers Central vacuole of cell Plasma membrane Secondary cell wall Primary cell wall Central vacuole of cell Middle lamella 1 µm Central vacuole Cytosol Plasma membrane Plant cell walls Figure 6.28 Plasmodesmata The Extracellular Matrix (ECM) of Animal Cells • Animal cells – Lack cell walls – Are covered by an elaborate matrix, the ECM • The ECM – Is made up of glycoproteins and other macromolecules EXTRACELLULAR FLUID Collagen A proteoglycan complex Polysaccharide molecule Carbohydrates Core protein Fibronectin Plasma membrane Integrin Figure 6.29 Integrins Microfilaments CYTOPLASM Proteoglycan molecule • Functions of the ECM include – Support – Adhesion – Movement – Regulation Intercellular Junctions Plants: Plasmodesmata • Plasmodesmata – Are channels that perforate plant cell walls Cell walls Interior of cell Interior of cell Figure 6.30 0.5 µm Plasmodesmata Plasma membranes Animals: Tight Junctions, Desmosomes, and Gap Junctions • In animals, there are three types of intercellular junctions – Tight junctions – Desmosomes – Gap junctions • Types of intercellular junctions in animals TIGHT JUNCTIONS Tight junction Tight junctions prevent fluid from moving across a layer of cells 0.5 µm At tight junctions, the membranes of neighboring cells are very tightly pressed against each other, bound together by specific proteins (purple). Forming continuous seals around the cells, tight junctions prevent leakage of extracellular fluid across A layer of epithelial cells. DESMOSOMES Desmosomes (also called anchoring junctions) function like rivets, fastening cells Together into strong sheets. Intermediate Filaments made of sturdy keratin proteins Anchor desmosomes in the cytoplasm. Tight junctions Intermediate filaments Desmosome Gap junctions Space between Plasma membranes cells of adjacent cells Figure 6.31 1 µm Extracellular matrix Gap junction 0.1 µm GAP JUNCTIONS Gap junctions (also called communicating junctions) provide cytoplasmic channels from one cell to an adjacent cell. Gap junctions consist of special membrane proteins that surround a pore through which ions, sugars, amino acids, and other small molecules may pass. Gap junctions are necessary for communication between cells in many types of tissues, including heart muscle and animal embryos. The Cell: A Living Unit Greater Than the Sum of Its Parts 5 µm • Cells rely on the integration of structures and organelles in order to function Figure 6.32