* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download - Horizon Discovery
Epigenetics in stem-cell differentiation wikipedia , lookup
Somatic cell nuclear transfer wikipedia , lookup
Therapeutic gene modulation wikipedia , lookup
Protein adsorption wikipedia , lookup
Chemical biology wikipedia , lookup
Cellular differentiation wikipedia , lookup
Transcriptional regulation wikipedia , lookup
Clinical neurochemistry wikipedia , lookup
Endogenous retrovirus wikipedia , lookup
Protein–protein interaction wikipedia , lookup
Gene regulatory network wikipedia , lookup
Point mutation wikipedia , lookup
Vectors in gene therapy wikipedia , lookup
Signal transduction wikipedia , lookup
Gene therapy of the human retina wikipedia , lookup
Paracrine signalling wikipedia , lookup
Expression vector wikipedia , lookup
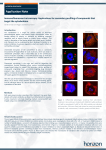
HORIZON DISCOVERY New Cellular Reporter Technologies Using X-MAN® Cell Models Paul Morrill1, Chris Torrance1, Christine Schofield1, Holly Astley1, Sue Rigby1, Tom Henley1, Claire Mahoney1, Paul Gonzales2, Bernardo Chavira2, Mei Cong3, Danette Daniels3, Hélène Benink3, Nancy Murphy3 and Jeff Kelly3 1Horizon Discovery Ltd, Cambridge, UK 2TGen Drug Development, Scottsdale, Arizona, USA Introduction X-MAN® (gene-X Mutant And Normal) cell lines provide genetically defined, patient-relevant, predictive in vitro models of genetic disease. We are further extending their application within targeted drug discovery with the development of new reporter disease models using our gene engineering technology (Figure 1). These models combine X-MAN® cell lines with the endogenous gene reporting capabilities in the form of NanoLuc® luciferase and HaloTag® reporter technologies. 3Promega Corporation, Madison, Wisconsin, USA X-MAN® HaloTag® Reporter Technology Mutated gene-X X-MAN® Nano-Glo®: NanoLuc® Luciferase Reporters NanoLuc® luciferase is a novel, small, bright luciferase reporter. It is one third the size and 100 times brighter than the most commonly used luciferase, firefly. These features enable the development of homogenous assays for primary and secondary screening, reporting on pathways via the endogenous promoter only, exploited using X-MAN® cell lines. NanoLuc® luciferase reporter lines allow transcriptional reporter output of endogenous promoter/response elements that drive NanoLuc® expression (Figure 2). Further applications include investigating changes in protein dynamics in response to the cellular environment or targeted therapies. (A) (C) (D) K-Ras HaloTag® WT Silver stain Figure 2. NanoLuc® luciferase detection at endogenous expression levels using HCT116 Hif1α NanoLuc® promoter fusion cell lines. (A) Correlation of cell number with luciferase signal. Even at low cell numbers a linear relationship is observed. (B) Correlation of concentration of Actinomycin D, a non-specific transcriptional inhibitor, with luciferase signal. A classical dose response is observed. t + 44 (0)1223 655580 f + 44 (0)1223 655581 e [email protected] w www.horizonbioproduction.com Horizon Discovery, 7100 Cambridge Research Park, Waterbeach, Cambridge, CB25 9TL, United Kingdom X-MAN-Glo® X-MAN® tumorigenic cell lines can be used for in vivo profiling studies. To further extend this capability, we have introduced a luciferase marker into X-MAN® cell lines, to allow non-invasive assessment of cancer progression in laboratory animals. Figure 6 shows images of the DLD-1 colon carcinoma line, which contains the heterozygous PI3Kα hinge domain mutation E545K, where the E545K mutation has been reverted to the wild type allele. Figure 6. DLD1 PI3Kα (E545K) knock-out xenografts. Firefly luciferase (Luc2) has been introduced into XMAN® cell lines to allow non-invasive radiance measurements. Images shown (left) are 11 days post infection. Other in vivo models containing luciferase include the SW48 K-Ras suite of mutations (including G12D, G12R and G12A). K-Ras HaloTag® WT cRaf 65-75 kDa Hif1α transcription Cell biomass Figure 5. SW48 mutant K-Ras HaloTag® variants. Confocal images of live cells labelled with HaloTag® ligand (red) and a DNA stain (green) show a similar pattern of localisation as for the wild type K-Ras HaloTag® fusion protein. Scale bars = 20 µm. (B) (B) Hif1α transcription (Luciferase RLU) Hif1α transcription (Luciferase RLU) (A) We have generated a suite of mutated K-Ras SW48 cell lines each with a HaloTag® reporter to investigate localisation and protein interactions between wild type and K-Ras G12C, G12V, G12D and G13D isogenic cell lines. These cell lines are identical except for the mutation of interest, which is expressed at the endogenous level. Figures 4 and 5 show analysis of the wild type and mutant cell lines by confocal microscopy and protein pull-down. SW48 K-Ras HaloTag® Mutant genotype: Isogenic background K-Ras G13D K-Ras G12V SW48 Parental Single Missense Mutation Figure 3. An overview of the HaloTag® reporter technology. Many functionalities are possible from a single fusion construct. SW48 HaloTag® Figure 1. An overview of the gene engineering technology. A virallymediated homologous recombination based technology that is highly efficient in performing gene-targeting in somatic human cell lines. Normal genotype: Isogenic background K-Ras G12D HaloTag® reporter technology encompasses a recombinant protein tag that allows flexibility between protein purification, expression and localization, protein interaction discovery, screening and other functional analysis (Figure 3). The technology is based on the formation of a covalent bond between the protein tag and synthetic ligands, and is designed to enable understanding of protein function in a cellular and biochemical environment. • X-MAN® Nano-Glo®: NanoLuc® luciferase reporters, with the key application in primary highthroughput screening and also further applications in protein degradation and stability studies • X-MAN® HaloTag®: Reporter disease models for secondary screening, with particular applications including protein purification, pull-downs and imaging studies • X-MAN-Glo®: For tagging existing X-MAN® cell models with luciferase for in vivo imaging applications Wild type gene-X K-Ras G12C WB against cRaf Figure 4. HaloTag® reporter images and pull-down. (A)-(C) Confocal experiments using SW48 wild type K-Ras HaloTag® cell lines show that K-Ras HaloTag® is present at the plasma membrane and in the cytoplasm. (A) KRas HaloTag® cells labelled with HaloTag® ligand show that the K-Ras HaloTag® fusion protein is localised to the plasma membrane and cytoplasm whereas cytoplasmic and nuclear staining is seen in SW48 parental cells transiently transfected with HaloTag® alone. (B) K-Ras HaloTag® cells labelled with HaloTag® ligand (red) and a DNA stain (green). (C) K-Ras HaloTag® cells labelled with HaloTag® ligand (red) and CellMask™ plasma membrane stain (green). (D) Pull-down experiments show enrichment of proteins for the K-Ras HaloTag® as compared to SW48 control cells. Enrichment of the known K-Ras interactor Raf is detected using an antibody against Raf. Scale bars = 20 µm. X-MAN® HaloTag® and Nano-Glo® lines in development A range of reporter lines are currently being generated and will be available by Q2/Q3 of 2012. Genotype Cell Line NanoLuc® promoter reporter Hif1α NanoLuc® protein reporter HaloTag® promoter reporter HaloTag® protein reporter NanoLuc® promoter reporter NRF2 NanoLuc® protein reporter HaloTag® promoter reporter HaloTag® protein reporter β-catenin NanoLuc® promoter reporter NanoLuc® protein reporter K-Ras HaloTag® promoter reporter p21 NanoLuc®promoter reporter cMyc NanoLuc® promoter reporter Gli1 NanoLuc® promoter reporter