* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download Epigenetics concerns changes in gene expression states that are
Genome evolution wikipedia , lookup
Epigenomics wikipedia , lookup
Cancer epigenetics wikipedia , lookup
Transgenerational epigenetic inheritance wikipedia , lookup
Primary transcript wikipedia , lookup
Gene expression profiling wikipedia , lookup
Epigenetics of neurodegenerative diseases wikipedia , lookup
Gene therapy of the human retina wikipedia , lookup
Y chromosome wikipedia , lookup
Epigenetics of diabetes Type 2 wikipedia , lookup
Microevolution wikipedia , lookup
Therapeutic gene modulation wikipedia , lookup
Behavioral epigenetics wikipedia , lookup
Non-coding RNA wikipedia , lookup
RNA silencing wikipedia , lookup
Genomic imprinting wikipedia , lookup
Genome (book) wikipedia , lookup
Designer baby wikipedia , lookup
Vectors in gene therapy wikipedia , lookup
Gene expression programming wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Epigenetics wikipedia , lookup
Site-specific recombinase technology wikipedia , lookup
Epigenetics in stem-cell differentiation wikipedia , lookup
Neocentromere wikipedia , lookup
Long non-coding RNA wikipedia , lookup
Nutriepigenomics wikipedia , lookup
Epigenetics of human development wikipedia , lookup
Polycomb Group Proteins and Cancer wikipedia , lookup
Mir-92 microRNA precursor family wikipedia , lookup
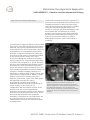
Mammalian Developmental Epigenetics U934/UMR3215 – Genetics and Developmental Biology Edith Heard Chef d'équipe [email protected] Tel: +33 1 56 24 66 91 Epigenetics concerns changes in gene expression states that are stable over rounds of cell division, but do not involve changes in the underlying DNA sequence of the organism. In female mammals, one of the two X chromosomes is transcriptionally silenced during early development to compensate for the double ‘dose’ of X-linked gene products in females (XX) when compared to males (XY). This process, known as X-chromosome inactivation (XCI), represents a paradigm for developmental epigenetics. A unique locus, the X-inactivation centre (Xic), initiates this process. The Xic produces a non-coding, regulatory RNA called Xist, which “coats” the X chromosome to be inactivated (Figure 1). We are interested in understanding the mechanisms by which X inactivation is initiated and maintained, via chromatin proteins, noncoding RNAs and DNA methylation. Understanding the epigenetics of X inactivation should provide important insights into diseases such as cancer, where deregulation of epigenetic states can play an important role. Figure 1 : X inactivation involves silencing of 1 of the 2 X chromosomes occurs during development Our group uses a combination of molecular genetics and cell biology approaches on embryos, embryonic stem cells and somatic cells. We are particularly interested in the roles that non-coding RNAs, chromatin changes and nuclear organisation might play in X INSTITUT CURIE, 20 rue d’Ulm, 75248 Paris Cedex 05, France | 1 Mammalian Developmental Epigenetics U934/UMR3215 – Genetics and Developmental Biology (left) via the non-coding XIst RNA (right). X Inactivation is triggered thanks to the lncRNA Xist, which coats the chromosome and induces silencing. We recently showed that the region that produces Xist (the X-inactivation centre) is organised into two topologically associating domains of sequence interactions (TADs) and uncovered a new level of chromosome folding in the mouse genome (Nora et al, 2012). Physical modelling has enabled us to predict the key regions of a chromatin fibre that allow it to fold in 3D (Giorgetti et al, 2014). Recently we have also investigated the degree to which autosomal loci show monoallelic expression across the genome. We found about 2% of loci show random monoallelic expression (RME) and that this is clonally heritable, similarly to Xinactivation. However RME is highly tissue and stage-specific implying that it allows a certain degree of plasticity in cellular expression patterns. Importantly, some of the RME genes we identified have specific roles in development and have been linked to autosomal dominant disorders (Gendrel et al, 2014). Finally, in the context of our collaboration with the medical section of the Institut Curie, we are investigating the epigenetic and genetic integrity of the inactive X chromosome in human breast cancer. Given the increasing realization that epigenetic instability is implicated on carcinogenesis, we are using the inactive X chromosome as a model system to assess this in different types of breast tumor. inactivation and gene expression in general. To this end we use multi-dimensional fluorescence imaging techniques, in both fixed and living cells, as well as genomic approaches to define chromatin and transcriptional states. Our studies on embryos and differentiating ES cells have shown that X inactivation is a highly dynamic process during early embryogenesis. We have also shown that the presence of two active X chromosomes leads to a delay in differentiation kinetics until X inactivation is achieved. Figure 2: Random monoallelic expression of several autosomal loci was revealed using allelespecific analyses in clonal neuronal progenitor cells and RNA FISH analyses in vivo in inbred mice (brain section of P6 stage shown with Sox2 biallelic expression and two monoallelic genes, Acyp2 and Cnrip1) INSTITUT CURIE, 20 rue d’Ulm, 75248 Paris Cedex 05, France | 2 Mammalian Developmental Epigenetics U934/UMR3215 – Genetics and Developmental Biology Key publications Year of publication 2017 Maud Borensztein, Laurène Syx, Katia Ancelin, Patricia Diabangouaya, Christel Picard, Tao Liu, Jun-Bin Liang, Ivaylo Vassilev, Rafael Galupa, Nicolas Servant, Emmanuel Barillot, Azim Surani, Chong-Jian Chen, Edith Heard (2017 Jan 31) Xist-dependent imprinted X inactivation and the early developmental consequences of its failure. Nature structural & molecular biology : DOI : 10.1038/nsmb.3365 Year of publication 2016 Luca Giorgetti, Bryan R Lajoie, Ava C Carter, Mikael Attia, Ye Zhan, Jin Xu, Chong Jian Chen, Noam Kaplan, Howard Y Chang, Edith Heard, Job Dekker (2016 Jul 21) Structural organization of the inactive X chromosome in the mouse. Nature : DOI : 10.1038/nature18589 Year of publication 2014 Luca Giorgetti, Rafael Galupa, Elphège P Nora, Tristan Piolot, France Lam, Job Dekker, Guido Tiana, Edith Heard (2014 May 1) Predictive polymer modeling reveals coupled fluctuations in chromosome conformation and transcription. Cell : 950-63 : DOI : 10.1016/j.cell.2014.03.025 Anne-Valerie Gendrel, Mikael Attia, Chong-Jian Chen, Patricia Diabangouaya, Nicolas Servant, Emmanuel Barillot, Edith Heard (2014 Feb 24) Developmental dynamics and disease potential of random monoallelic gene expression. Developmental cell : 366-80 : DOI : 10.1016/j.devcel.2014.01.016 Edda G Schulz, Johannes Meisig, Tomonori Nakamura, Ikuhiro Okamoto, Anja Sieber, Christel Picard, Maud Borensztein, Mitinori Saitou, Nils Blüthgen, Edith Heard (2014 Feb 6) The two active X chromosomes in female ESCs block exit from the pluripotent state by modulating the ESC signaling network. Cell stem cell : 203-16 : DOI : 10.1016/j.stem.2013.11.022 Year of publication 2011 Elphège P Nora, Bryan R Lajoie, Edda G Schulz, Luca Giorgetti, Ikuhiro Okamoto, Nicolas Servant, INSTITUT CURIE, 20 rue d’Ulm, 75248 Paris Cedex 05, France | 3 Mammalian Developmental Epigenetics U934/UMR3215 – Genetics and Developmental Biology Tristan Piolot, Nynke L van Berkum, Johannes Meisig, John Sedat, Joost Gribnau, Emmanuel Barillot, Nils Blüthgen, Job Dekker, Edith Heard (2011 Oct 3) Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature : 381-5 : DOI : 10.1038/nature11049 INSTITUT CURIE, 20 rue d’Ulm, 75248 Paris Cedex 05, France | 4