* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download Histones
Gel electrophoresis of nucleic acids wikipedia , lookup
Cancer epigenetics wikipedia , lookup
United Kingdom National DNA Database wikipedia , lookup
Site-specific recombinase technology wikipedia , lookup
Human genome wikipedia , lookup
DNA damage theory of aging wikipedia , lookup
No-SCAR (Scarless Cas9 Assisted Recombineering) Genome Editing wikipedia , lookup
DNA vaccination wikipedia , lookup
Hybrid (biology) wikipedia , lookup
Molecular cloning wikipedia , lookup
Epigenetics of human development wikipedia , lookup
Point mutation wikipedia , lookup
Therapeutic gene modulation wikipedia , lookup
Nucleic acid analogue wikipedia , lookup
Nucleic acid double helix wikipedia , lookup
Genealogical DNA test wikipedia , lookup
Primary transcript wikipedia , lookup
Non-coding DNA wikipedia , lookup
Cre-Lox recombination wikipedia , lookup
Cell-free fetal DNA wikipedia , lookup
Epigenomics wikipedia , lookup
Genomic library wikipedia , lookup
Genome (book) wikipedia , lookup
Polycomb Group Proteins and Cancer wikipedia , lookup
Skewed X-inactivation wikipedia , lookup
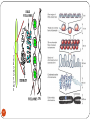
Deoxyribozyme wikipedia , lookup
Designer baby wikipedia , lookup
Vectors in gene therapy wikipedia , lookup
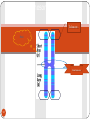
History of genetic engineering wikipedia , lookup
Microevolution wikipedia , lookup
Extrachromosomal DNA wikipedia , lookup
DNA supercoil wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Comparative genomic hybridization wikipedia , lookup
Y chromosome wikipedia , lookup
X-inactivation wikipedia , lookup
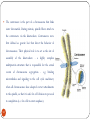
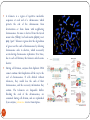
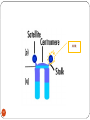
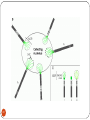
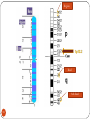
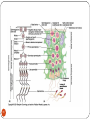
Chapter 8 Chromosome and chromatin structure :)امام صادق (ع .برترین عبادت مداومت نمودن بر تفکر درباره خداوند و قدرت اوست 1 Dr. S Hosseini-Asl; Radiology; ARUMS; 1392 DNA bases 2 Nucleotide 3 dNTPs 4 5 6 ds DNA 7 DNA vs. RNA 8 Histones Histones are highly alkaline proteins found in eukaryotic cell nuclei that package and order the DNA into structural units called nucleosomes. They are the chief protein components of chromatin, acting as spools around which DNA winds, and play a role in gene regulation. Without histones, the unwound DNA in chromosomes would be very long (a length to width ratio of more than 10 million to one in human DNA). For example, each human cell has about 1.8 meters of DNA, but wound on the histones it has about 90 micrometers (0.09 mm) of chromatin, which, when duplicated and condensed during mitosis, result in about 120 micrometers of chromosomes. 9 10 11 12 Human Chromosome Telomeres Petit Centromere 13 Centromere The centromere is the part of a chromosome that links sister chromatids. During mitosis, spindle fibers attach to the centromere via the kinetochore. Centromeres were first defined as genetic loci that direct the behavior of chromosomes. Their physical role is to act as the site of assembly of the kinetochore - a highly complex multiprotein structure that is responsible for the actual events of chromosome segregation - e.g. binding microtubules and signaling to the cell cycle machinery when all chromosomes have adopted correct attachments to the spindle, so that it is safe for cell division to proceed to completion (i.e. for cells to enter anaphase). 14 15 Telomere A telomere is a region of repetitive nucleotide sequences at each end of a chromosome which protects the end of the chromosome from deterioration or from fusion with neighboring chromosomes. Its name is derived from the Greek nouns telos (τέλος) 'end' and merοs (μέρος, root: μερ-) 'part.' Telomere regions deter the degradation of genes near the ends of chromosomes by allowing chromosome ends to shorten, which necessarily occurs during chromosome replication. Over time, due to each cell division, the telomere ends become shorter. During cell division, enzymes that duplicate DNA cannot continue their duplication all the way to the end of chromosomes. If cells divided without telomeres, they would lose the ends of their chromosomes, and the necessary information they contain. The telomeres are disposable buffers blocking the ends of the chromosomes, are consumed during cell division, and are replenished by an enzyme, telomerase reverse transcriptase. Telomere shortening and aging 16 Chromosome types Metacentric p=q Submetacentric p<q Acrocentric p<<q Telocentric p=0 17 Human Chromosome groups A: 1-3, MC (2=SMC) B: 4,5, SMC C: 6-12,X , SMC D: 13-15 , AC E: 16-18 , SMC F: 19,20 , MC G: 21,22,Y , AC 18 Acrocentric chromosomes NOR 19 Nucleolus NOR: Nucleolus Organizing Region 20 Techniques for chromosome studies 1. Routine cytogenetic 21 Solid staining 22 G-banding G-banding is a technique used in cytogenetics to produce a visible karyotype by staining condensed chromosomes. It is useful for identifying genetic diseases through the photographic representation of the entire chromosome complement. The metaphase chromosomes are treated with trypsin (to partially digest the chromosome) and stained with Giemsa. Dark bands that take up the stain are strongly A,T rich (gene poor). The reverse of G-bands is obtained in R-banding. Banding can be used to identify chromosomal abnormalities, such as translocations, because there is a unique pattern of light and dark bands for each chromosome. It is difficult to identify and group chromosomes based on simple staining because the uniform color of the structures makes it difficult to differentiate between the different chromosomes. Therefore, techniques like G-banding were developed that made "bands" appear on the chromosomes. These bands were the same in appearance on the homologous chromosomes, thus, identification became easier and more accurate.The acid-saline-Giemsa protocol reveals G-bands. 23 R-, Q-, C-banding Different chromosomal staining techniques reveal variations in chromosome structure. Cytogeneticists use these patterns to recognize the differences between chromosomes and enable them to link different disease phenotypes to chromosomal abnormalities. Giemsa banding (a), Q-banding (b), R-banding (c) and Cbanding (d) 24 Techniques for chromosome studies 2. Molecular cytogenetic 25 ISH (In Situ Hybridization) 26 In situ hybridization (ISH) is a type of hybridization that uses a labeled complementary DNA or RNA strand (i.e., probe) to localize a specific DNA or RNA sequence in a portion or section of tissue (in situ), or, if the tissue is small enough (e.g. plant seeds, Drosophila embryos), in the entire tissue (whole mount ISH), in cells and in circulating tumor cells (CTCs). This is distinct from immunohistochemistry, which usually localizes proteins in tissue sections. DNA ISH can be used to determine the structure of chromosomes. FISH (Fluorescent ISH) 27 SKY (Spectral Karyotyping) Spectral karyotyping is a molecular cytogenetic technique used to simultaneously visualize all the pairs of chromosomes in an organism in different colors. Fluorescently labeled probes for each chromosome are made by labeling chromosome-specific DNA with different fluorophores. Because there are a limited number of spectrally-distinct fluorophores, a combinatorial labeling method is used to generate many different colors. Spectral differences generated by combinatorial labeling are captured and analyzed by using an interferometer attached to a fluorescence microscope. Image processing software then assigns a pseudo color to each spectrally different combination, allowing the visualization of the individually colored chromosomes. 28 SKY 29 CGH (Comparative Genomic Hybridization) 30 Comparative genomic hybridization (CGH) or Chromosomal Microarray Analysis (CMA) is a molecularcytogenetic method for the analysis of copy number changes (gains/losses) in the DNA content of a given subject's DNA and often in tumor cells. CGH will detect only unbalanced chromosomal changes. Structural chromosome aberrations such as balanced reciprocal translocations or inversions cannot be detected, as they do not change the copy number. Region 1p13.2 Band Sub-band 31 32 33 34 Spermatogenesis 35 36 Oogenesis 37