* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download Control, Genomes and Environment
Non-coding DNA wikipedia , lookup
Transposable element wikipedia , lookup
Point mutation wikipedia , lookup
Public health genomics wikipedia , lookup
Oncogenomics wikipedia , lookup
History of RNA biology wikipedia , lookup
X-inactivation wikipedia , lookup
Genetic engineering wikipedia , lookup
Messenger RNA wikipedia , lookup
Epigenetics of neurodegenerative diseases wikipedia , lookup
Short interspersed nuclear elements (SINEs) wikipedia , lookup
Vectors in gene therapy wikipedia , lookup
Pathogenomics wikipedia , lookup
Quantitative trait locus wikipedia , lookup
Therapeutic gene modulation wikipedia , lookup
Gene expression programming wikipedia , lookup
Nutriepigenomics wikipedia , lookup
RNA silencing wikipedia , lookup
Long non-coding RNA wikipedia , lookup
Non-coding RNA wikipedia , lookup
Essential gene wikipedia , lookup
Epitranscriptome wikipedia , lookup
Site-specific recombinase technology wikipedia , lookup
RNA interference wikipedia , lookup
Primary transcript wikipedia , lookup
Polycomb Group Proteins and Cancer wikipedia , lookup
Genomic imprinting wikipedia , lookup
Genome evolution wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Designer baby wikipedia , lookup
History of genetic engineering wikipedia , lookup
Genome (book) wikipedia , lookup
Microevolution wikipedia , lookup
Ridge (biology) wikipedia , lookup
Biology and consumer behaviour wikipedia , lookup
Minimal genome wikipedia , lookup
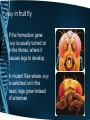
Control, Genomes and Environment Cellular Control – homeobox genes and apoptosis Learning Outcomes • explain that the genes that control development of body plans are similar in plants, animals and fungi, with reference to homeobox sequences (HSW1) • outline how apoptosis (programmed cell death) can act as a mechanism to change body plans Controlling development All organisms begin life as a single cell. This cell divides and the new cells produced start to differentiate and specialize. ‘Switching on’ the expression of a gene or keeping it switched off determines the development of features. Many organisms contain similar genes that control development of body plans. For example groups of genes called the homeobox genes play an important role in the development of many multicellular organisms. Homeobox genes • Homeobox genes determine how an organism’s body develops as it grows from a zygote into a complete organism. • They determine the organism’s body plan • These sequences are highly conserved, which implies that their activity is fundamental to the development of an organism • Homeobox genes have been discovered in animals, plants and fungi Homeobox genes The genome of the fruit fly contains one ‘set’ or cluster of homeobox genes. These control development, including the polarity of the embryo, polarity of each segment and the identity of each segment. Homeobox genes code for transcriptional factors. These regulate the expression of other genes important in development. Mutations in homeobox genes can cause changes in the body plan. For example a mutation in the gene controlling leg placement can cause legs to grow where the antennae are normally found. Homeobox genes Homeobox genes are present in the genomes of most organisms. They control development of body parts in similar ways. There is little variation in many regions of the homeobox genes in different organisms. This suggests that these have been highly conserved throughout evolutionary history. They are thought to be especially important to the basic development of organisms. Homologous homeobox genes • These are the sequences of 60 amino acids in the proteins coded for by the homeobox genes Antp in a fruit fly and HoxB7 in a mouse. • All animals have homologous homeobox genes – they are recognisably similar What do homeobox genes do? • Homeobox genes code for the production of transcription factors (homeodomain) • These proteins can bind to a particular region of DNA and activate or repress the gene – A single homeobox gene can switch on a whole collection of other genes, regulating gene expression • This allows the correct development of the body plan. Genes and Body plans • Drosophila melanogaster a.k.a. fruit fly • Body is divided into – Head – Thorax – abdomen Genetic control of Drosophila development • Development is mediated by homeobox genes – Maternal effect genes determine the embryo’s polarity e.g. anterior (head) & posterior (tail / abdomen) – Segmentation genes determine polarity of each segment – Homeotic selector genes identify and direct the development of each segment • Two groups exist, that control development of (i) head + thorax segments and (ii) thorax + abdomen segments. Drosophila thorax • The Thorax of the fruit fly is split into 3 segments – T1 – a pair of legs – T2 – a pair of legs and a pair of wings – T3 – a pair of legs and a pair of halteres Ubx in fruit fly • A homeobox gene called Ubx stops the formation of wings in T3. • A mutation in both copies of Ubx, wings grow in T3 instead of halteres. Ubx Mutant Ubx Antp in fruit fly • If the homeobox gene Antp is usually turned on in the thorax, where it causes legs to develop. • In mutant flies where Antp is switched on in the head, legs grow instead of antennae Hox Clusters • Hox clusters are aggregations of homeobox genes and are found in all animals. • Examples • Nematodes have one Hox cluster • Fruit flies have 2 Hox clusters • Vertebrates have 4 Hox clusters Read p114-5 in purple book • Add any information to your notes made on homeobox genes and answer q on p115. • Include the first example of the interference of homeobox functioning in humans. Homeobox genes in humans • Effect of thalidomide in embryo development – Homeobox genes HoxA11 and HoxD11 switch on genes that cause forelimb development. – The drug thalidomide affected the behaviour of these homeobox genes at a critical stage in embryonic development. Apoptosis – programmed cell death • Cell death as part of normal development • Happens in stages • Complete the sheet Apoptosis: • programmed cell death. • Ensure you are clear about the stages and the example of syndactyly • Purple book 116-7, Green book p105 to help. Plenary • Suggest how one gene may inhibit the action of another. (3 marks) • • • • • • codes for inhibitor; protein; blocks transcription (of allele coding for pigment); ref to, regulator / promoter; blocks enzyme (producing pigment); AVP; e.g. detail Spec Check Homework • Read the scientific article (start of) which will give you more detail about the potential role of apoptosis in cancer treatment. • Find out about siRNA Learning Outcomes • explain that the genes that control development of body plans are similar in plants, animals and fungi, with reference to homeobox sequences (HSW1) • outline how apoptosis (programmed cell death) can act as a mechanism to change body plans Control, Genomes and Environment Cellular Control – Translational control siRNA Translational control • Small interfering RNA (siRNA) are short pieces of double-stranded RNA that interferes with the expression of a specific gene. • The siRNA is complementary to the mRNA produced during transcription. • It binds to the mRNA, it is chopped into pieces and cannot combine with the ribosome. • Thus it cannot be translated. siRNA inhibits translation of mRNA and turns genes OFF siRNA unwinds. Now single stranded Complementary base pairing between siRNA and target mRNA Enzyme 1 breaks up dsRNA making siRNA molecules Enzyme 2 cuts mRNA into small sections stopping it from being translated Exam Practice • Q7, Q8, Q14 in mrsmillers F215 document Answers • Q7 (a) (i) mRNA leaves nucleus; ora mRNA, translated / used to make, protein; DNA, transcribed / used to make, mRNA; mRNA short-term / DNA (long-term) store; 2 max • (ii) siRNA smaller / fewer nucleotides / only matches part of gene; ora siRNA double-stranded; ora 2 Answers • Q8 (b) (complementary) base-pairing; hydrogen bonding; between purines and pyrimidines; A with U; R A with T C with G; ref to 2 or 3 bonds (correct context); 3 max • 8. (i) (CCR5 / macrophages) (siRNAs continue to work) in long-lived cells; only one treatment needed for macrophages / CCR5; (siRNAs diluted) as lymphocytes divide; ora repeat treatments needed for, lymphocytes / CD4; 2 • (ii) (CCR5) because no essential function in body / absence not a problem; 1 Answers • • • • • • • • • • 14. (i) RNA(i) combines with mRNA; e.g. of base pairing (but not T) A-U / G-C; stops translation; ref to stops mRNA combining with ribosomes; stops protein synthesis; max 3 (ii) chemicals / enzymes in, mouth / toothpaste / bacteria; denature / degrade, RNA; RNA not normally taken up by bacterial cells; short life of RNA; RNA not replicated in bacteria when bacteria reproduce; Answers • (ii) chemicals / enzymes in, mouth / toothpaste / bacteria; • denature / degrade, RNA; • RNA not normally taken up by bacterial cells; • short life of RNA; • RNA not replicated in bacteria when bacteria reproduce; • toothpaste in mouth only for short time; • AVP; • AVP; e.g. washed away by saliva max 2