* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download HANDOUT: CH 18 pt 1 Study
History of RNA biology wikipedia , lookup
Gene desert wikipedia , lookup
Epigenomics wikipedia , lookup
Gene therapy wikipedia , lookup
Epigenetics of depression wikipedia , lookup
Cancer epigenetics wikipedia , lookup
Epigenetics in stem-cell differentiation wikipedia , lookup
Short interspersed nuclear elements (SINEs) wikipedia , lookup
Gene therapy of the human retina wikipedia , lookup
Vectors in gene therapy wikipedia , lookup
Protein moonlighting wikipedia , lookup
Biology and consumer behaviour wikipedia , lookup
Genetic engineering wikipedia , lookup
RNA interference wikipedia , lookup
Metabolic network modelling wikipedia , lookup
Genome (book) wikipedia , lookup
Genome evolution wikipedia , lookup
Gene nomenclature wikipedia , lookup
History of genetic engineering wikipedia , lookup
Point mutation wikipedia , lookup
Polycomb Group Proteins and Cancer wikipedia , lookup
Epigenetics of neurodegenerative diseases wikipedia , lookup
RNA silencing wikipedia , lookup
Gene expression programming wikipedia , lookup
Designer baby wikipedia , lookup
Epigenetics of diabetes Type 2 wikipedia , lookup
Site-specific recombinase technology wikipedia , lookup
Epigenetics in learning and memory wikipedia , lookup
Microevolution wikipedia , lookup
Long non-coding RNA wikipedia , lookup
Epigenetics of human development wikipedia , lookup
Epitranscriptome wikipedia , lookup
Gene expression profiling wikipedia , lookup
Primary transcript wikipedia , lookup
Mir-92 microRNA precursor family wikipedia , lookup
Therapeutic gene modulation wikipedia , lookup
Nutriepigenomics wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
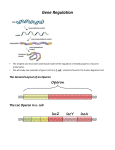
WLHS / AP Bio / Monson Name Date Per CHAPTER 18 STUDY QUESTIONS, part 1 – Regulation of Gene Expression: Prokaryotes vs. Eukaryotes (p. 351-366) 1) What are the two levels within which metabolic control can occur in bacteria? 2) What is the advantage of feedback inhibition? In what kinds of pathways is this mechanism typically found? 3) What is the key advantage of grouping genes of related function (e.g. the genes encoding all of the enzymes in the tryptophan synthesis pathway) into one transcription unit? 4) Label each component / product of the trp operon shown below - (see fig. 18.3a) (environmental / cellular conditions: tryptophan absent, repressor inactive, operon on). 5) How does a repressor work? What kind of gene encodes a repressor protein? 6) How is the trp repressor protein an allosteric protein? 7) Briefly, what is the key difference between a repressible operon and an inducible operon? 8) Why is the trp operon considered REPRESSIBLE? 9) Why is the lac operon considered INDUCIBLE? 10) Why are repressible enzymes generally associated with anabolic pathways and how is this an advantage to the organism? 11) Why are inducible enzymes generally associated with catabolic pathways and how is this an advantage to the organism? 12) How does positive gene regulation work? 13) Explain why it is said that the lac operon “has both an on-off switch and a volume control.” 14) In general, what is the effect of histone acetylation and DNA methylation on gene expression? 15) What is meant by alternative RNA splicing? What is the significance of alternative RNA splicing? 16) Summarize how miRNAs and siRNAs interact with and affect mRNA. 17) What is the evolutionary significance of small non-coding RNAs (ncRNAs)? (HINT: see “Evolution” section on page 366) 18) Label the figure below showing the stages of gene expression that can be regulated in eukaryotic cells. (HINT: see fig. 18.6 on page 356)