* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Ch 10 Taxonomy and Classification
Metagenomics wikipedia , lookup
Horizontal gene transfer wikipedia , lookup
Bacterial cell structure wikipedia , lookup
Bacterial morphological plasticity wikipedia , lookup
Human microbiota wikipedia , lookup
Sarcocystis wikipedia , lookup
Triclocarban wikipedia , lookup
Community fingerprinting wikipedia , lookup
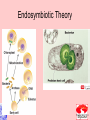
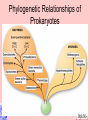
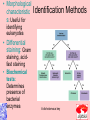
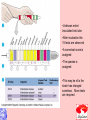
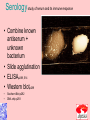
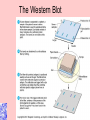
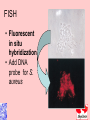
Taxonomy and Classification Classification of Microbes Taxonomy • Science of Classification of organisms • Hopes to show relationships among organisms • Is a way to provide universal identification of an organism • Why do we care things are related? Q&A • Pneumocystis jirovecii was thought to be a protozoan until DNA analysis showed it is a fungus. Why does it matter whether an organism is classified as a protozoan or a fungus? Phylogeny or Systematics • Shows evolutionary relationships and history among organisms • Some obtained from fossil record • Most bacteria use rRNA sequencing or some other sequence information • A goal is to identify all organisms by 2025 Hierarchy • Evolutionary relationships • Species are groups that interbreed (have productive sex) • How this goes • • • • • • • • • Species Genus Family Order Class Division Phylum Kingdom (1969) Domain (80’s) The 5 Kingdoms based on nutrient procurement • Plantae – Multicellular photoautotrophs • Animalia – ingestive • Fungi – absorptive • Protozoa – Mostly singe celled • Prokaryotes • Which of these are microbes? The 3 domains • Eukarya – Plants, animals fungi and protists • Bacteria – (with peptidoglycan) • Archaea – With unusual cell walls, and membreanes The Three-Domain System Figure 10.1 The Three-Domain System Table 10.1 Endosymbiotic Theory Figures 10.2, 10.3 Table 10.2 Scientific Nomenclature • Binomial genus and specific epithet (species). Is used world wide • Is always underlined • Rules for naming are set by international committee’s – International Code of Zoological Momenclature – International Code of Botanical Nomenclature – Bacteriological Code and Bergey’s Manual Scientific Names Scientific Binomial Source of Genus Name Source of Specific Epithet Klebsiella pneumoniae Honors Edwin Klebs The disease Pfiesteria piscicida Honors Lois Pfiester Disease in fish Salmonella typhimurium Honors Daniel Salmon Stupor (typh-) in mice (muri-) Streptococcus pyogenes Chains of cells (strepto-) Forms pus (pyo-) Penicillium chrysogenum Tuftlike (penicill-) Produces a yellow (chryso-) pigment Trypanosoma cruzi Corkscrew-like (trypano-, borer; soma-, body) Honors Oswaldo Cruz Species Definition • Eukaryotic species: – A group of closely related organisms that breed among themselves • Prokaryotic species: – – – – A population of cells with similar characteristics Clone: Population of cells derived from a single cell Strain: Genetically different cells within a clone Culture: grown in the lab • Viral species: – Population of viruses with similar characteristics that occupies a particular ecological niche Is it as easy to classify Microbes as it is Macrobes? • How to classify • What do we have to look at? Identifying Bacteria Applications, p. 283 Phylogenetic Relationships of Prokaryotes Figure 10.6 Of more than 2600 species identified so far • Only about 250 or 10% are pathogens Classification and Identification • Classification: Placing organisms in groups of related species. Lists of characteristics of known organisms. • Identification: Matching characteristics of an “unknown” organism to lists of known organisms. – Clinical lab identification Bergey’s Manual of Systematic Bacteriology • • • • • • • • Morphological characteristics Presence of various enzymes Serological tests Phage typing Fatty acid profiles DNA finger printing Sequence of ribosomal RNA Is still very difficult • Morphological characteristic s: Useful for Identification Methods identifying eukaryotes • Differential staining: Gram staining, acidfast staining • Biochemical tests: Determines presence of bacterial enzymes A dichotomous key Figure 10.8 A clinical microbiology lab report form Figure 10.7 Identifying a Gram – Negative, Oxidase – Negative Rod Figure 10.8 •Unknown enteri inoculated into tube •After incubation the 15 tests are observed •A numerical score is assigned •The species is assigned •This may be of is the strain has changed somehow. More tests are required • Design a rapid test for a Staphylococcus aureus. 10-14 Figure 6.10 Serology study of serum and its immune responce • Combine known antiserum + unknown bacterium • Slide agglutination • ELISAp288, 514 • Western blotp289 • • Southern Blot p292 DNA chip p293 Figure 10.10 • Strains with different antigens are called – Serotypes – Serovars – biovars DNA Hybridization Phage Typing Determining a strains suceptability to certain phage or bacterial viruses Figure 10.13 The Western Blot Flow Cytometry • Uses differences in electrical conductivity between species • Fluorescence of some species • Cells selectively stained with antibody plus fluorescent dye Figure 18.12 A typical dichotomous key See appendix H in your lab book Genetics • DNA base composition – Guanine + cytosine moles% (GC) • DNA fingerprinting – Electrophoresis of restriction enzyme digests • rRNA sequencing • Polymerase Chain Reaction (PCR)p251 Figure 10.14 Nucleic Acid Hybridization: DNA Chip DNA chip Technolog y Figure 10.17 Differentiate between classificaiton and identification Figure 10.5 FISH • Fluorescent in situ hybridization • Add DNA probe for S. aureus Figure 10.18 Differentiate between strain and species? Classification of viruses? • Not currently placed in Domains or Kingdoms • Why? • Species are usually a population of viruses with similar characteristics that occupies a particular ecological niche. Dichotomous keys are used for identification of organisms Cladograms show phylogenetic relationships among organisms Differential staining • Name examples The gram stain Using Bergies manual • Used to Identify bacteria not classify • Features that are used to differentiate various organism often have little to do with arranging the orgs in taxonomic groups 4 major groups • Domain Bacteria – Gram-negative Eubacteria that have cell walls. Proteobacteria – Non proteobacteria Gram negative bacteria – Gram positive Eubacteria that have cell walls • Domain archaeobacteria Some groups by identificaiton • Spirochetes – Genus Borrelia, Leptospira, Treponema • Aerobic/microaerophilic, motikle, helical/vibroid gram negative bacteria – Geneus Camphylobacter • Gram negative aerobic/microaerophilic rods and cocci – Genus Agrobacterium, Alcaligenes, Pseudomonas • Facultatively anaerobic gram negative rods – Genus Enterobater, Escherichia, Klebsiella, Serratia, Shigella, Yersinia, Eikenella • Gram positive Cocci – Genus lactococcus, streptococcus, Staphylococcus • Endospore forming gram positive rods and cocci – Genus bacillus, clostridium • Regular, nonsporing gram positive rods – Genus Lactobacillus, listeria