* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Lecture 13
Triclocarban wikipedia , lookup
Metagenomics wikipedia , lookup
Bacterial cell structure wikipedia , lookup
Magnetotactic bacteria wikipedia , lookup
Human microbiota wikipedia , lookup
Molecular mimicry wikipedia , lookup
Virus quantification wikipedia , lookup
Marine microorganism wikipedia , lookup
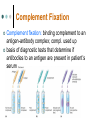
Clinical Microbiology Lecture 13 Bio 3124 Identification of pathogens is critical Use appropriate treatments • Antibiotics don’t work on all bacteria • Many bacteria are now drug-resistant • Proper choice of antibiotics necessary Required for proper prognosis • Streptococcal pharyngitis might appear like a mild infection • Could cause serious heart, kidney complications Track spread of disease • Allows faster treatment of others infected • Allows identification of cause of infection Clinical Microbiology, Specimens Clinical microbiology isolate and identify microbes from clinical specimens rapidly Clinical specimen human material tested to determine the presence or absence of specific microbes specimen should: represent diseased area Sufficient quantity to do a variety of diagnostic tests collected aseptically to avoid contamination obtained prior to administration of antimicrobial forwarded promptly and properly to a clinical lab Clinical Techniques Definitive identification relies on: Microscopy: Morphological assessment, fluorescence microscopy for specific detection Biochemical techniques, require growing pathogen Immunologic tests: use of antibodies, Elisa, Agglutination test, complement fixation, immunoprecipitation based tests eg. radial immunodiffusion, double diffusion Molecular techniques: PCR, QPCR, Ribotyping, RFLP also phage typing Microscopy wet-mount, heat-fixed, or chemically fixed specimens can be examined choice of microscopy depends on pathogen Morphological, Gram reaction, spore bearing e.g., dark-field microscopy • detection of spirochetes in skin lesions associated with syphilis e.g., fluorescence microscopy stains often used Simple stains, Gram stain and acid fast stain (Zeil-Neelsen) for mycobacteria Immunofluorescence microscopy fluorophores are exposed to UV, violet, or blue light to make them fluoresce coupled to antibody molecules without changing antibody’s ability to bind a specific antigen can be used as direct fluorescent-antibody (FA) assay or indirect fluorescent-antibody (IFA) assay Direct and indirect Immunofluorescence detection Cytomegalovirs infected cells Herpes simplex infected cells Growth and Biochemical Characteristics Viruses Sample used to infect cells in tissue culture identified by: immunodiagnostic tests molecular methods replication in culture detected by: Polio induced CPE cytopathic effects • morphological changes in host cells Syncytium hemadsorption • binding of red blood cells to surface of infected cells (hemagglutinin producing viruses) Hemadsorption hPIV3 induced cell fusion Biochemical Identification Bacteria most bacteria: culturing in growth media • can provide preliminary information about biochemical nature of bacterium additional biochemical tests used following isolation some bacteria are not routinely cultured rickettsias, chlamydiae, and mycoplasmas identified with special stains, immunologic tests, or molecular methods such as PCR Biochemical tests Examples of biochemical tests Biochemical properties represent genetic relatedness Database of biochemical capabilities Can be used to identify bacteria Growing on different substrates as sole carbon source Biochemical signature of test organism Compare with database to find the best match See Flowcharts (algorithms) for ID’ing schemes Lac- = dark blue Hemin NAD NAD+Hemin H. Influenza requires NAD and hemin Lac+ = yellow Oxidase test N. meningitidis is has cyt C Identification scheme for G+ bacteria Identification scheme for Gbacteria Rapid Methods of Identification manual biochemical systems mechanized/automated systems: e.g., API 20 E system Biolog phenotypic arrays immunologic systems Reference book: Bergey's manual of determinative bacteriology / [edited by] John G. Holt et al., Baltimore : Williams & Wilkins, c1994 API 20E system Checks for 20 metabolic markers and generates codes to match known bacteria Biolog phenotypic identification array More than 2500 bacterial, fungi and yeast species Based on colorimetric detection of growth Use of a redox dye coupled to ETC 95 metabolic markers Rapid 4-16 hours Computer based database match Accuracy Biolog Inc website Each well contains one carbon source growth results in color change Bacteriophage Typing based on specificity of phage surface molecules for host cell receptors Narrow host range for a collection of phages can be used to typify the hosts Phagovars collection of strains sensitive to certain collection of phage types Molecular Methods Nucleic acid-based detection methods • Ribotyping • Diagnostic PCR • Probe hybridization (RFLP analysis) Analysis of proteins: PAGE and Western Ribotyping To identify bacterial genera based on high level of 16S rRNA conservation among bacteria PCR amplification of rRNA genes or fragments Sequence of amplified DNA compared with those in the National Center for Biotechnology (NCBI) Strain is determined on the basis of sequence homology Diagnostic PCR Amplifies small fragment of DNA Allows detection of tiny numbers of bacteria Size of fragment can indicate species, strain Clostridium botulinum toxin genes Samples Restriction analysis can further indicate strain Single nucleotide differences affect ability to be cut by restriction enzymes Real-Time Quantitative PCR Detection of slow growing viruses, latent infections RT-PCR Reverse Transcriptase makes cDNA from RNA followed by PCR qRT-PCR: quantitative “real-time” PCR Quenched fluorescent probe to amplified DNA • Probe is degraded as amplification occurs • Separates quencher from fluorophore Measure appearance of fluorescence • Faster the gain, the more template present • Indicates more viral RNA or DNA in sample Animation: Real-time PCR Genomic fingerprint: RFLP analysis RFLP (restriction fragment length polymorphism) • Genomic DNA restriction • Electrophoesis • Chemical denaturation • Southern transfer: transfer of ssDNA onto nylon membranes • Probe hybridization: short complementary DNA labelled with 32P or tagged with an enzyme eg Alkaline phosphatase • Detection: chromogenic reaction or by X-ray autoradiography - Related strains show similar RFLP patterns Animation: Southern blot and RFLP analysis Immunologic Techniques detection of antigens or antibodies in specimens especially useful when culture methods are unavailable or impractical use of immunological techniques has many advantages easy to use rapid reaction endpoints sensitive and specific Agglutination agglutinates visible immune complexes formed by cross-linking cells with antibodies eg., Agglutination of S. thyphi by serum from infected patient (Widal Test) Can be used to titre the serum antibodies for a pathogen titer = reciprocal of highest dilution positive for agglutination Complement Fixation Complement fixation: binding complement to an antigen-antibody complex; compl. used up basis of diagnostic tests that determine if antibodies to an antigen are present in patient’s serum Animation: Complement fixation Enzyme-Linked Immunosorbent Assay Done in two ways: Direct Indirect Direct: directly detecting antigens in a sample also called antigen capture ELISA Pathogen specific Ab immobilized Patient serum passed Secondary antibody detects binds to captured Ag Enzyme-Linked Immunosorbent Assay Indirect: detecting antibodies in a patients serum interpreted as indirect evidence for exposure to a pathogen Known antigen immobilized Primary Ab detects Ag Enz-linked secondary Ab detects primary Ab Can be quantified Animation: Direct and Indirect ELISA Immunoblotting (Western Blot) procedure proteins separated by SDS-PAGE proteins transferred to nitrocellulose sheets protein bands visualized with enzyme-tagged antibodies Examples: distinguish microbes diagnostic tests Extra slides if you like to seek for more Monoclonal Antibodies (MAB) and fluorescence microscopy produced by hybridoma cells recognize a single epitope fluorescently-labeled mABs used diagnostically • technique has replaced use of polyclonal antisera for culture confirmation Click to read more about hybridoma technology