* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download 090923-eNMR-Barcelona
Survey
Document related concepts
Transcript
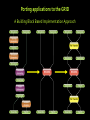
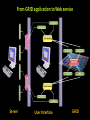
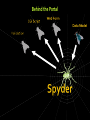
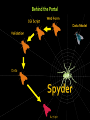
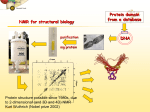
Structural Biology on the GRID Dr. Tsjerk A. Wassenaar Biomolecular NMR - Utrecht University (NL) [email protected] 2007: e-Infrastructure grant contract no. 213010 - e-NMR: Streamline and automate NMR structure determination 2009: 2nd largest VO in life sciences 236 dedicated CPU (473 kSI2K) 2.87 Tb storage 3290 CPU at supporting sites 35.25 Tb of storage 8 web portals 115 registered users ~450 server requests ~375k jobs ~110 normalized CPU years Structural Biology on the GRID Function and Mechanism of Action Design of Experiments Understand Effects of Mutations Drug Design NMR: Nuclear Magnetic Resonance NMR data processing NMR data processing • Many programs • Expertise required • Demanding in terms of CPU and data storage eNMR: Objectives Develop and provide integrated protocols and interoperability of NMR applications Offer easy access web interfaces for NMR data processing Exploit GRID technology for computationally demanding structure calculations Lower the access level to GRID technology in life sciences eNMR: Challenges Porting applications to the GRID Interoperability of applications Automated data processing and error handling User access Security Porting applications to the GRID Call for simple and effective application control: From UI or SE To UI or SE Porting applications to the GRID A Building Block Based Implementation Approach From GRID application to Web service Server User Interface GRID Security For services offer usage, don’t offer access Users can use eNMR services either with a certificate loaded or Users register with a certificate to obtain a login and password (jobs can then be run with a robot certificate) Users can only run prefabricated jobs with limited control http://www.enmr.eu/webportal The eNMR Web Services HADDOCK: Structure Prediction of Protein-Protein Complexes CYANA: NMR Structure Calculations XPLOR-NIH: NMR Structure Calculations csRosetta: NMR Structure Calculations TALOS+: Backbone Torsion Angle Prediction MDD NMR: Processing of non-uniformly sampled nD spectra CCPNmr: Format Conversion eNMR Database The HADDOCK Web Portal A protocol for every user: Easy Interface: Minimal choice, simple to use Expert Interface: Many options for tailored runs Guru Interface: Almost full control File upload: Complete control Use of the server requires registration The service is GRID enabled when run through eNMR http://www.enmr.eu/ http://haddock.chem.uu.nl/enmr Behind the Portal Behind the Portal Challenges From simple services to complete workflows Making services available as web services (WSDL) Critical Assessment of Structure Determination by NMR Nature Methods 6, 625-626 (2009) Meet up to business requirements, regarding security, provenance and accounting Beat BioMed as the largest VO in life sciences ;) Conclusions eNMR has become established, including infrastructure, services and a growing user base. eNMR has yielded innovative solutions to bridge user needs, user friendliness and GRID resources eNMR is lowering the barriers between life sciences and NMR and between life sciences and GRID computing Utrecht University Thank you for your attention! Dr. Tsjerk A. Wassenaar Biomolecular NMR - Utrecht University (NL) [email protected] http://skriptic.wordpress.com/