* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download SWTMedicalInformatics
Data center wikipedia , lookup
Data analysis wikipedia , lookup
Entity–attribute–value model wikipedia , lookup
Data vault modeling wikipedia , lookup
3D optical data storage wikipedia , lookup
Web analytics wikipedia , lookup
Information privacy law wikipedia , lookup
Business intelligence wikipedia , lookup
Open data in the United Kingdom wikipedia , lookup
Relational model wikipedia , lookup
Database model wikipedia , lookup
Web Ontology Language wikipedia , lookup
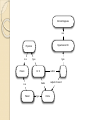
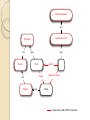
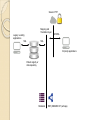
Semantic Web Technologies: A Paradigm for Medical Informatics Chimezie Ogbuji (Owner, Metacognition LLC.) http://metacognition.info/presentations/SWTMedicalInformatics.pdf http://metacognition.info/presentations/SWTMedicalInformatics.ppt Who I am Circa 2001: Introduced to web standards and Semantic Web technologies 2003-2011: Lead architect of CCF in-house clinical repository project 2006-2011: Member representative of CCF in World-wide Web Consortium (W3C) ◦ Editor of various standards and Semantic Web Health Care and Life Sciences Interest Group chair 2011-2012: Senior Research Associate at CWRU Center for Clinical Investigations 2012-current: Started business providing resource and data management software for home healthcare agencies (Metacognition LLC) Medical Informatics Challenges Semantic interoperability ◦ Exchange of data with common meaning between sender and receiver Most of the intended benefits of HIT depend on interoperability between systems Difficulties integrating patient record systems with other information resources are among the major issues hampering their effectiveness ◦ Interoperability is a major goal for meaningful use of Electronic Health Records (EHR) Rodrigues et al. 2013; Kadry et al. 2010; Shortliffe and Cimino, 2006 Requirements and Solutions Semantic interoperability requires: ◦ Structured data ◦ A common controlled vocabulary Solutions emphasize the meaning of data rather than how they are structured ◦ “Semantic” paradigms Registries and Research DBs Patient registries and clinical research repositories capture data elements in a uniform manner The structure of the underlying data needs to be able to evolve along with the investigations they support Thus, schema extensibility is important Querying Interfaces Standardized interfaces for querying facilitate: ◦ Accessibility to clinical information systems ◦ Distributed querying of data from where they reside Requires: ◦ Semantically-equivalent data structures Alternatively, data are centralized in data warehouses Austin et al. 2007, “Implementation of a query interface for a generic record server” Biomedical Ontologies Ontologies are artifacts that conceptualize a domain as a taxonomy of classes and constraints on relationships between their members Represented in a particular formalism Increasingly adopted as a foundation for the next generation of biomedical vocabularies Construction involves representing a domain of interest independent of behavior of applications using an ontology Important means towards achieving semantic interoperability Biomedical Ontology Communities Prominent examples of adoption by life science and healthcare terminology communities: ◦ The Open Biological and Biomedical Ontologies (OBO) Foundry ◦ Gene Ontology (GO) ◦ National Center for Biomedical Ontology (NCBO) Bioportal ◦ International Health Terminology Standards Development Organization (IHTSDO) Semantic Web and Technologies The Semantic Web is a vision of how the existing infrastructure of the World-wide Web (WWW) can be extended such that machines can interpret the meaning of data on it Semantic Web technologies are the standards and technologies that have been developed to achieve the vision An Analogy (Technological) singularity is a theoretical moment when artificial intelligence (AI) will have progressed to a greater-thanhuman intelligence Despite remaining in the realm of science fiction, it has motivated many useful developments along the way ◦ The use of ontologies for knowledge representation and IBM Watson capabilities, for example Background: Graphs Graphs are data structures comprising nodes and edges that connect them The edges can be directional Either the nodes, the edges, or both can be labeled The labels provide meaning to the graphs (edge labels in particular) Node edge Node Resource Description Framework The Resource Description Framework (RDF) is a graph-based knowledge representation language for describing resources It’s edges are directional and both nodes and edges are labeled It uses Universal Resource Identifiers (URI) for labeling Foundation for Semantic Web technologies RDF: Continued The edges are statements (triples) that go from a subject to an object Some objects are text values Some subjects and objects can be left unlabeled (Blank nodes) ◦ Anonymous resources: not important to label them uniquely The URI of the edge is the predicate Predicates used together for a common purpose are a vocabulary Dr. X author treats subject of record Chime full name "Chimezie Ogbuji" Subject: Dr. X (a URI) Object: Chime Predicate: treats Vocabulary: ◦ treats, subject of record, author, and full name RDF vocabularies How meaning is interpreted from an RDF graph There are vocabularies that constrain how predicates are used ◦ Want a sense of treats where the subject is a clinician and the object is a patient There is a predicate relating resources to the classes they are a member of (type) There are vocabularies that define constraints on class hierarchies These comprise a basic RDF Schema (RDFS) language Represented as an RDF graph Clinical Diagnosis is a Hypertension DX Physician is a Person type type Dr. X subject of record treats is a Patient author type Chime Ontologies for RDF The Ontology Web Language (OWL) is used to describe ontologies for RDF graphs More sophisticated constraints than RDFS Commonly expressed as an RDF graph Defines the meaning of RDF statements through constraints: ◦ On their predicates ◦ On the classes the resources they relate belong to Clinical Diagnosis is a Hypertension DX Physician is a Person type type Dr. X subject of record treats is a Patient author type Chime Governed by OWL/ RDFS for domain OWL Formats Most common format for describing ontologies Distribution format of ontologies in the NCBO BioPortal SNOMED CT distributions include an OWL representation ◦ RDF graphs can describe medical content in a SNOMED CT-compliant way through the use of this vocabulary Validation and Deduction OWL is based on a formal, mathematical logic that can be used for validating the structure of an ontology and RDF data that conform to it (consistency checking) Used to deduce additional RDF statements implied by the meaning of a given RDF graph (logical inference) Logical reasoners are used for this Inference Can infer anatomical location from SNOMED CT definitions Hypertension DX Systemic circulatory system structure type type finding site Hypertension DX <-> 1201005 / “Benign essential hypertension (disorder)” Querying RDF Graphs SPARQL is the official query language for RDF graphs Comparable to relational query languages ◦ Primary difference: it queries RDF triples, whereas SQL queries tables of arbitrary dimensions Includes various web protocols for querying RDF graphs Foundation of SPARQL is the triple pattern (?clinician, treats, ?patient) ◦ ?clinician and ?patient are variables (like a wildcard) Which physicians have given essential hypertension diagnoses and to whom? Hypertension DX type ?physician treats author ?dx subject of record (?physician, author, ?dx) (?physician, treats, ?patient) (?dx, subject of record, ?patient) (?dx, type, Hypertension DX) ?patient ?physician ?patient ?dx Dr. X Chime … SPARQL over Relational Data Most common implementations convert SPARQL to SQL and evaluate over: ◦ a relational databases designed for RDF storage ◦ an existing relational database There are products for both approaches Former requires native storage of RDF ◦ Relational structure doesn’t change even as RDF vocabulary does (schema extensibility) Elliot et al. 2009, “A Complete Translation from SPARQL into Efficient SQL” SPARQL over Existing Relation Data “Virtual RDF view” ◦ Translation to SQL follows a given mapping from existing relational structures to an RDF vocabulary ◦ Allows non-disruptive evolution of existing systems ◦ Well-suited as a standard querying interface over clinical data repositories ◦ They can be queried as SPARQL, securely over encrypted HTTP Secure HTTP Mapping and Translation layer Legacy / existing applications SQL SPARQL SQL 3rd party applications Patient registry or data repository Relational RDF (SNOMED CT perhaps) Example: Cleveland Clinic (SemanticDB) Content repository and data production system released in Jan. 2008 80 million (native) RDF statements ◦ Uses vocabulary from a patient record OWL ontology for the registry Based on ◦ Existing registry of heart surgery and CV interventions ◦ 200,000 patient records ◦ Generating over 100 publications per year Pierce et al. 2012, “SemanticDB: A Semantic Web Infrastructure for Clinical Research and Quality Reporting” Cohort Identification Interface developed in conjunction with Cycorp Leverage their logical reasoning system (Cyc) ◦ Identifies cohorts using natural language (NL) sentence fragments ◦ Converts fragments to SPARQL ◦ SPARQL is evaluated against RDF store Example: Mayo Clinic (MCLSS) Mayo Clinic Life Sciences System (MCLSS) ◦ Effort to represent Mayo Clinic EHR data as RDF graphs ◦ Patient demographics, diagnoses, procedures, lab results, and free-text notes ◦ Goal was to wrap MCLSS relational database and expose as read-only, query-able RDF graphs that conform to standard ontologies ◦ Virtual RDF view Pathak et al. 2012, "Using Semantic Web Technologies for Cohort Identification from Electronic Health Records for Clinical Research" Example: Mayo Clinic (CEM) Clinical Element Model (CEM) ◦ Represents logical structure of data in EHR ◦ Goal: translate CEM definitions into OWL and patient (instance) data into conformant RDF ◦ Use tools (logical reasoners) to check semantic consistency of the ontology, instance data, and to extract new knowledge via deduction ◦ Instance data validation: correct number of linked components, value within data range, existence of units, etc. Tao et al. 2012, ”A semantic-web oriented representation of the clinical element model for secondary use of electronic health records data" Summary Schema extensibility ◦ Use of RDF Semantic Interoperability ◦ Domain modeling using OWL and RDFS Standardized query interfaces ◦ Querying over SPARQL Incremental, non-disruptive adoption ◦ Virtual RDF views Main challenge: highly disruptive innovation