* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download NMR Spectroscopy
Survey
Document related concepts
Transcript
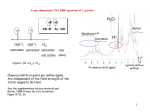
NMR Spectroscopy Part II. Signals of NMR Free Induction Decay (FID) • FID represents the time-domain response of the spin system following application of an radio-frequency pulse. • With one magnetization at w0, receiver coil would see exponentially decaying signal. This decay is due to relaxation. Fourier Transform The Fourier transform relates the time-domain f(t) data with the frequency-domain f(w) data. Fourier Transform Fourier Transform NMR line shape Lorentzian line y AW 2 W 4x0 x 2 A amplitude W half-line width 2 Resolution Definition For signals in frequency domain it is the deviation of the peak line-shape from standard Lorentzian peak. For time domain signal, it is the deviation of FID from exponential decay. Resolution of NMR peaks is represented by the half-height width in Hz. Resolution Resolution-digital resolution Resolution Measurement half-height width: 10~15% solution of 0-dichlorobenzene (ODCB) in acetone Line-shape: Chloroform in acetone Resolution Factors affect resolution Relaxation process of the observed nucleus Stability of B0 (shimming and deuterium locking) Probe (sample coil should be very close to the sample) Sample properties and its conditions Sensitivity Definition signal to noise-ratio A s / n 2.5 N pp A: height of the chosen peak Npp : peak to peak noise Sensitivity Measurement 1H 0.1% ethyl benzene in deuterochloroform 13C ASTM, mixture of 60% by volume deuterobenzene 31P 15N and dioxan or 10% ethyl benzene in chloroform 1% trimehylphosphite in deuterobenzene 19F 90% dimethylformamide in deutero-dimethylsulphoxide 0.1% trifluoroethanol in deuteroacetone 2H, 17O tap water Sensitivity Factors affect sensitivity Probe: tuning, matching, size Dynamic range and ADC resolution Solubility of the sample in the chosen solvent Spectral Parameters Chemical Shift Caused by the magnetic shielding of the nuclei by their surroundings. d-values give the position of the signal relative to a reference compound signal. Spin-spin Coupling The interaction between neighboring nuclear dipoles leads to a fine structure. The strength of this interaction is defined as spinspin coupling constant J. Intensity of the signal Chemical Shift Origin of chemical shift Beff B0 sB0 1 s B0 s shielding constant 1 s B0 Beff 2 2 ' Chemically non-equivalent nuclei are shielded to different extents and give separate resonance signals in the spectrum Chemical Shift Chemical Shift d – scale or abscissa scale B0 1 s 1 1 2 B0 1 s 2 2 2 B0 s 2 s 1 2 1 2 2 1 s 2 s1 1 Chemical shift parameter d s 2 s 1 10 6 Chemical Shift 6 d 10 observing frequency Shielding s CH3Br < CH2Br2 < CH3Br < TMS d CHBr 3 90 MHz spectrum 614 90 10 6 10 6 6.82 (ppm) Abscissa Scale Chemical Shift d is dimensionless expressed as the relative shift in parts per million ( ppm ). d is independent of the magnetic field d of proton 0 ~ 13 ppm d of carbon-13 0 ~ 220 ppm d of F-19 0 ~ 800 ppm d of P-31 0 ~ 300 ppm Chemical Shift s local s dia s local para s N s R s e si Charge density Neighboring group Anisotropy Ring current Electric field effect Intermolecular interaction (H-bonding & solvent) Chemical Shift – anisotropy of neighboring group sN 1 3r 3 4 // 1 cos2 susceptibility r distance to the dipole’s center Differential shielding of HA and HB in the dipolar field of a magnetically anisotropic neighboring group Chemical Shift – anisotropy of neighboring group d~2.88 d~9-10 • Electronegative groups are "deshielding" and tend to move NMR signals from neighboring protons further "downfield" (to higher ppm values). • Protons on oxygen or nitrogen have highly variable chemical shifts which are sensitive to concentration, solvent, temperature, etc. • The -system of alkenes, aromatic compounds and carbonyls strongly deshield attached protons and move them "downfield" to higher ppm values. •Electronegative groups are "deshielding" and tend to move NMR signals from attached carbons further "downfield" (to higher ppm values). •The -system of alkenes, aromatic compounds and carbonyls strongly deshield C nuclei and move them "downfield" to higher ppm values. •Carbonyl carbons are strongly deshielded and occur at very high ppm values. Within this group, carboxylic acids and esters tend to have the smaller values, while ketones and aldehydes have values 200. Ring Current The ring current is induced form the delocalized electron in a magnetic field and generates an additional magnetic field. In the center of the arene ring this induced field in in the opposite direction t the external magnetic field. Ring Current -- example Spin-spin coupling Spin-spin coupling AX system AX2 system Spin-spin coupling AX3 system Multiplicity Rule Multiplicity M (number of lines in a multiplet) M = 2n I +1 n equivalent neighbor nuclei I spin number For I= ½ M=n+1 Example AX4 AX4 system I=1; n=3 Order of Spectrum Zero order spectrum only singlet First order spectrum >> J Higher order spectrum ~ J AMX system Spin-spin coupling Hybridization of the atoms Bond angles and torsional angles Bond lengths Neighboring -bond Effects of neighboring electron lone-pairs Substituent effect JH-H and Chemical Structure Geminal couplings 2J (usually <0) H-C-H bond angle hybridization of the carbon atom substituents Geminal couplings J 2 bond angle Geminal couplings J 2 Substituent Effects Effect of Neighboring -electrons Vicinal couplings JH-H 3 Torsional or dihedral angles Substituents HC-CH distance H-C-C bond angle Vicinal couplings JH-H 3 3 Karplus curves 1 3 1 3 J 2 J g Jt 3 3 dihedral angles Chemical Shift of amino acid http://bouman.chem.georgeto wn.edu/nmr/interaction/chems hf.htm Chemical Shift Prediction Automated Protein Chemical Shift Prediction http://www.bmrb.wisc.edu:8999/shifty.html BMRB NMR-STAR Atom Table Generator for Amino Acid Chemical Shift Assignments http://www.bmrb.wisc.edu/elec_dep/gen_aa.html http://bouman.chem.georgetown.edu/nmr/interaction/chemshf.htm Example 1