* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download slides
Survey
Document related concepts
Transcript
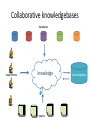
Provenance in a Collaborative Bio-database RAASWiki Donald Dunbar & Jon Manning Queen’s Medical Research Institute University of Edinburgh Use Cases for Provenance April 20th 2009 Provenance in Bio-databases including RAASWiki Donald Dunbar & Jon Manning Queen’s Medical Research Institute University of Edinburgh Use Cases for Provenance April 20th 2009 Plan bio-databases RAASWiki collaborative knowledgebases provenance Biological databases • Sequences – Ensembl, Entrez • Structure – PDB • Expression – GEO, ArrayExpress – Function – Gene Ontology – Interaction – MINT, BIND, KEGG – ‘Warehouses’ – GeneCards, IUPHAR – Literature – Pubmed How do they handle provenance? Ensembl produces genome databases for vertebrates and other eukaryotic species, and makes this information freely available online. ‘Gene’ ID histories (with stable ID) Evidence for gene predictions Links to other databases (eg Uniprot) How do they handle provenance? The PDB archive contains information about experimentally-determined structures of proteins, nucleic acids, and complex assemblies. Primary citation History: deposition and last update Raw data and protocols How do they handle provenance? Gene Expression Omnibus: a gene expression/molecular abundance repository supporting MIAME compliant data submissions, and a curated, online resource for gene expression data browsing, query and retrieval. Standards compliance (protocols, data…) Links within database (microarrays, protocols) Raw data and protocols How do they handle provenance? The Gene Ontology project provides a controlled vocabulary to describe gene and gene product attributes in any organism. Evidence for gene annotation (experimental, computational) Links to original publications No versioning, just updates How do they handle provenance? PubMed is a free search engine for accessing the MEDLINE database of citations, abstracts and some full text articles on life sciences and biomedical topics. Original source material, authors, abstracts Unique Pubmed ID (used by other databases) Continual updates (new papers), occasional retractions How do they handle provenance? GeneCards® is a searchable, integrated database of human genes that provides concise genomic, proteomic, transcriptomic, genetic and functional information on all known and predicted human genes. Lots of data from other databases IDs/keys from sources Lots of data integration based on IDs How do they handle provenance? The IUPHAR database (IUPHAR-DB) integrates peer-reviewed pharmacological, chemical, genetic, functional and anatomical information on GPCRs, ligand-gated ion channels and voltage-gated-like ion channel subunits encoded by the human, rat and mouse genomes. Curated by experts Original sources plus curation provenance Suggested citations Newer developments WikiGenes is the first wiki system to combine the collaborative and largely altruistic possibilities of wikis with explicit authorship. In view of the extraordinary success of Wikipedia there remains no doubt about the potential of collaborative publishing, yet its adoption in science has been limited. Here I discuss a dynamic collaborative knowledge base for the life sciences that provides authors with due credit and that can evolve via continual revision and traditional peer review into a rigorous scientific tool. but…. RAASWiki Important biology - hypertension RAASWiki is a knowledgebase of information on the renin-angiotensin-aldosterone system. While much of the seed data were derived from pre-existing databases such as KEGG and OMIM, supplementary data are included not easily available through such resources. This includes short textual reports on the genes involved, and more experimentally-oriented information such as animal models. Automatic seeding of database (BioKB) Collaborative editing (Wiki based, useful functionality) Genes, publications, animal models, datasets… RAASWiki – provenance Seeded data tagged with source database and date Edits are tagged with editor and date Comments are tagged: name and date Wiki functionality allows versioning and roll back Identifiers for source databases preserves provenance ‘Crowd wisdom’ will hopefully unsure good quality RAASWiki – provenance issues How much detail (each edit, granularity, versions)? Who will use provenance data? Different focus depending on data (who, when, confidence) How much should we rely on sources for provenance? Annotation & comments v changing data Public v private data Likely to become a big issue What provenance to we need? Example: Gene expression in a transgenic animal gene expression measurements gene annotation where, when public databases which identifiers integration when, what, how output from machine how processing what and how did we select genes data mining … What provenance to we need? Example: Curated gene database database links curation contributor, date curator input source, identifiers, dates development verify, add, delete, modify archive versions, dates Curated database schema & interface changes Collaborative knowledgebases databases experiments knowledge papers knowledgebase Collaborative knowledgebase provenance issues Confidence in data Tracking data to its (real) source Published papers do not contain all information When is something (knowledge) finished Citing of knowlegebase records Linking between knowledgebase records Some sort of dynamic publication Conclusions • • • • • In biology provenance is a mixed bag We use mainly static databases Usually source is clear but not much else RAASWiki contains static and curated data We have implemented a very rudimentary provenance scheme • Collaborative knowledgebases will need to address provenance in new ways