* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download SGKB
Survey
Document related concepts
Transcript
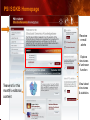
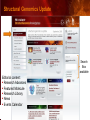
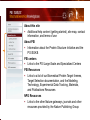
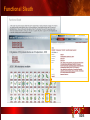
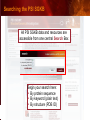
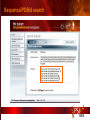
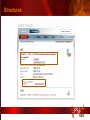
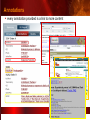
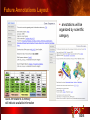
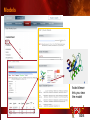
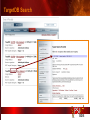
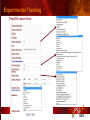
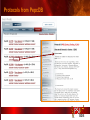
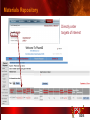
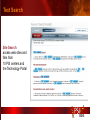
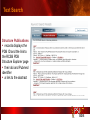
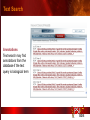
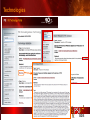
PSI Structural Genomics Knowledgebase Helen M. Berman, Rutgers University EMBO Practical Course Section: Searching Structure Databases September 26, 2008 Knowledgebase Knowledgebase Vision The PSI Structural Genomics Knowledgebase (PSI SGKB) will turn the products of the PSI effort into major advances in knowledge that can be used to understand living systems and human disease. It will be a key resource for the advancement of biology, biochemistry, functional genomics, pharmacology, bioinformatics, chemistry, education and clinical medicine. SGKB Knowledgebase Goals To provide a “marketplace of ideas” that connects protein sequence information to 3D structures and homology models enhances functional annotations provides access to new experimental protocols and materials To kick start and enable advancements in structural genomics by communicating and providing visibility and accessibility of information and technology advances of the PSI through presentation and discussion of the most provocative challenges with the general community by fostering community collaborations SGKB PSI SGKB features Database searchable by sequence, text, and PDB ID Search results include aggregate reports and inventories Links to PSI projects, external resources, and publications SG Gateway with Nature delivers featured articles, PSI news and events, featured molecules and technologies, molecules of unknown function and broader SG content Notification to public about recently solved PSI structures or new editorial content SGKB Scope Experimental Tracking Target Selection Materials Genomic Based Target Selection Isolation, Expression, Purification, Crystallization Data Collection Structure Determination PDB Deposition & Release Models Annotations Publications Technology Metrics To capture, make accessible, and highlight elements of the highthroughput pipelines for use by various scientific communities To leverage such information through the generation of molecular models and functional annotation SGKB Knowledgebase Users Biologists Biochemists Functional Genomicists Pharmacologists Bioinformatics Chemists Clinical Researchers and Physicians Teachers and Students SGKB A Tour of the PSI SGKB PSI SGKB Homepage Receive e-mail alerts Explore structures of unknown function Teasers for this month’s editorial content View latest structures & statistics 1 SGKB Structural Genomics Update Search Box available Editorial content: Research Advances Featured Molecule Research Library News Events Calendar 1 SGKB About this site Additional help content (getting started), site map, contact information, and terms of use About PSI Information about the Protein Structure Initiative and the PSI SGKB PSI centers Links to the PSI Large-Scale and Specialized Centers PSI Resources Links to a list of our Biomedical Protein Target themes, Target Selection documentation, and the Modeling, Technology, Experimental Data Tracking, Materials, and Publications Resources NPG Resources Links to the other Nature gateways, journals and other resources provided by the Nature Publishing Group 1 SGKB E-alerts: Receive news of PSI SGKB updates by email or RSS feed Updates to editorial content (monthly) Newly released structures (weekly) Functional Sleuth: explore protein structures solved by the PSI whose functions are unknown Latest PSI statistics Provides current tallies of structures solved View detailed reports of which structures have solved by the PSI (“Metrics”) View the latest structures solved by the PSI 1 SGKB Functional Sleuth SGKB Metrics PSI-2 Summary Statistics Updated Sept 5, 2008 I.1.A Number of novel experimental PSI-2 structures 1219 I.1.B Number of distinct experimental PSI-2 structures non-redundant sequences 1714 I.1.D Total number of experimental PSI-2 structures 1933 I.1.E Numbers of experimentally determined distinct residues 385746 Numbers of experimentally determined novel residues 268559 I.2.J Number of experimental structures of human proteins 71 I.2.K Number of experimental structures of eukaryotic proteins 206 I.2.M Number of experimental structures of membrane proteins 10 I.2.N Number of experimental structures determined at the atomic level using x-ray crystallography 1753 Number of experimental structures determined at the atomic level using NMR methods 180 novel structures - structures with less than 30% sequence identity to an existing structure at the time of PDB deposition distinct proteins - structures with non-redundant sequences less 1 than 98% sequence identity SGKB See latest structures… SGKB Searching the PSI SGKB Searching the PSI SGKB SGKB Searching the PSI SGKB All PSI SGKB data and resources are accessible from one central Search Box Begin your search here: By protein sequence By keyword (plain text) By structure (PDB ID) 1 SGKB Sequence/PDBid search Available structures of proteins with similar/identical amino acid sequences Any structural and functional properties (annotation) determined from these protein structures Available theoretical/homology models created with amino acid sequences similar to your query Any information about similar protein sequences (targets) studied by the PSI structural genomics efforts The protocols used during those PSI research efforts Ordering information to obtain DNA clone materials, if available. SGKB Sequence/PDBid search SGKB Structures In the Structures tab, experiment and reference information about the structure is displayed: View matching sequence alignment and sequence identity Link to RCSB PDB’s Structure Explorer to learn more about the structure View information about chemical substrates in the experiment (bound ligands and substrates) Download the 3D atomic coordinates for the molecule If published, connect to its citation and abstract at PubMed. SGKB Structures SGKB Annotations Genomic features: gene identifier, name and synonyms, operon/regulon mappings from databases Protein sequence features: amino acid sequence, taxonomy & phylogeny, isoforms, single nucleotide polymorphisms, post-translational modifications, and sequence families. Structure features: secondary structure, oligomeric state, structure and functional domains, DNA binding motifs, sites of interaction Ligands: information about bound ligands Functional/Biochemical classifications: enzyme class, substrate specificity and catalysis, epitope mapping, cellular location, organ location Protein Networks and Biological Systems: enzymatic pathways and networks information Literature: synonyms for protein names, links to PubMed by database identifier and related text and authors Information from more than 50 external annotation resources SGKB Annotations every annotation provided is a link to more content SGKB Future Annotations Layout annotations will be organized by scientific category Quick Annotations Summary will indicate available information SGKB Models In the Models tab, a list of the homology models available from the integrated Protein Models Portal are displayed view the structural model, and interact with it in a Java window (AstexViewer) download the model’s atomic coordinates view predicted domain annotations from databases such as InterPro view sequence/domain annotations related to the template structure, such as SCOP and CATH SGKB Models AstexViewer lets you view the model SGKB Experimental Data Tracking TargetDB contains worldwide structural genomics protein target information. Search by sequence, Target ID, project site, status, update date, protein name, and source organism Links to other sequence databases, domain databases, other structural genomics centers, and the RCSB PDB Download target data Target statistics summary PepcDB contains all the functionality of TargetDB plus Experimental protocols Detailed status history of experimental trials Information on failed experiments SGKB TargetDB Search SGKB Experimental Tracking PepcDB search form SGKB Protocols from PepcDB SGKB Materials Repository Directly order targets of interest SGKB Text Search With a plain text search, find information from: PSI Center web pages Publications resource Technology resource Annotation database SGKB Text Search Site Search access web sites and files from 10 PSI centers and the Technology Portal SGKB Text Search Structure Publications records displays the PDB ID and the link to the RCSB PDB Structure Explorer page their doi and Pubmed identifier a link to the abstract SGKB Text Search Annotations Text search may find annotations from the database if the text query is biological term SGKB Text Search Methodology Publications their doi and Pubmed identifier a link to the abstract SGKB Technology Module PSI Centers are actively developing technologies and methodologies for all aspects of the structure determination pipeline Genomic Based Target Selection Isolation, Expression, Purification, Crystallization Data Collection Structure Determination PDB Deposition & Release Publications Functional Annotation SGKB Technologies SGKB Publications to Date SGKB Acknowledgements Access Information http://kb.psi-structuralgenomics.org KB Group Wendy Tao Raship Shah James Chun Margaret Gabanyi Tom Oldfield John Westbrook PSI Resources Nature Matthew Day Andrei Kouranov (Exp. Data Tracking) Boyana Konforti Torsten Schwede (Models) Paul Adams (Technology) KB Steering Committee Josh La Baer (Materials) Chair, Eaton Lattman Wladek Minor (Publications) SGKB