* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download Court Lab Blurb for Sackler web site
Survey
Document related concepts
Transcript
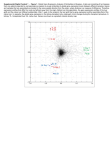
Members: Suwagmani Hazarika, M.Sc., Research Assistant 2000 - BSc (honors), Zoology, Banaras Hindu University, Banaras, India. 2002 - MSc, Biotechnology, India Institute of Technology, Roorke, India. Carrie Mosher, Ph.D., Postdoctoral Fellow 1998 - BSc, Biological and Environment Sciences, University of Notre Dame, Notre Dame, IN. 2002 - MSc, Pharmaceutical Chemistry, Idaho State University, Pocatello, ID. 2008 - PhD, Medicinal Chemistry, University of Washington, Seattle, WA. Awewura Kwara, MB.ChB., MPH&TM, NIH-K23 Award Trainee 1992 - MB.ChB., Medical School, University of Ghana, Accra, Ghana. 1999 - Residency, Cook County Hospital, Chicago, IL. 2001 - MPH&TM, School of Public Health and Tropical Medicine, Tulane University, New Orleans, LA. 2004 - Assistant Professor of Medicine, Brown University, Providence, RI. Laboratory of Comparative and Molecular Pharmacogenomics The long-term goal of the research being conducted in our laboratory is personalized therapeutics - the safe and effective treatment of an individual’s disease using the right dose of the right medicine tailored specifically for that patient. Unfortunately, many drugs used today are characterized by high interindividual variability in therapeutic response and adverse effects necessitating a trial and error approach using escalating doses and different classes of drugs to achieve a successful outcome. Our research involves identifying significant causes of this variability, particularly those resulting from differences in the genetic makeup of an individual (i.e. pharmacogenomics). The ultimate outcome of this work will be the development of rational drug treatment algorithms based on a patient’s genotype, predictive biomarkers, demographics, disease state, as well as other coadministered drugs. Drug glucuronidation enzymology and pharmacogenomics The UDP-glucuronosyltransferases (UGTs) are an unappreciated and understudied superfamily of drug metabolizing enzymes that metabolize and inactivate drugs by conjugation with glucuronic acid (i.e. glucuronidation). UGTs are second only to the cytochromes P450 (CYPs) in terms of the number of clinically important drugs that are substrates for these enzymes. Primary pharmaceutical substrates for the UGTs that have been extensively studied in our lab include acetaminophen (Tylenol), azidothymidine (AZT), morphine, mycophenolic acid, oxazepam, and lorazepam. Published and ongoing studies involve establishing the responsible UGT isoforms for each of these (and other) drugs, as well as identifying genetic variants that predict increased or decreased rates of drug glucuronidation in a patient. Fig. 1. Variability in glucuronidation of probes specific for UGT1A1, 1A4, 1A6, 1A9, 2B7 and 2B15 measured in the same bank of human liver microsomes. Also shown are activities for midazolam 1’-hydroxylation and chlorzoxazone 6hydroxylation, as probes for CYP3A and CYP2E1. Data for each activity are given relative to the mean activity for all livers. Also shown are the coefficients of variation (CV%) for each activity. Acetaminophen-induced acute liver failure Acetaminophen is one of the most commonly used drugs in the world, but is also the leading cause of acute liver failure in the United States and Europe. While many cases result from intentional overdose (i.e. suicide attempts), as many cases are the result of therapeutic use of this drug for chronic pain relief. We are currently conducting studies using cell and tissue-based model systems, human volunteers, and DNA collected from patients that developed acetaminopheninduced acute liver failure, to identify genotypes predictive of altered acetaminophen metabolism and increased risk for developing liver injury. Such genotypes would be used to identify individuals that should avoid or perhaps minimize the use of acetaminophen. Fig. 2. Etiology of acute liver failure in the United States as determined by the Acute Liver Failure Study Group multicenter trial (courtesy Will Lee, UTSW, 2009). Almost half of the cases are the result of acetaminophen ingestion, and of those approximately half were taking acetaminophen for therapeutic purposes. HIV drug pharmacogenomics and TB drug interactions Antiretroviral drug combination therapies have had a substantial impact on reducing the morbidity and mortality associated with HIV infection. However, certain highly effective antiretroviral drugs, such as efavirenz, demonstrate high interindividual variability in blood levels resulting in either adverse neurological effects (high levels) or potential virologic failure (low levels). Research in our laboratory and others have established that polymorphisms in the gene encoding CYP2B6 are largely responsible for high variability in efavirenz metabolism. In collaboration with researchers at Brown University, University of North Carolina, and the University of Ghana, we are identifying genetic variants in CYP2B6 and other genes that are predictive of slow as well as fast metabolism of efavirenz using DNA samples from a cohort of HIV-infected Ghanaians. We are also exploring the molecular basis for variable drug-drug interactions between efavirenz and the drugs used to treat tuberculosis (TB), a common comorbid infection in HIV-infected patients. The ultimate goal is to develop rational therapeutic protocols based on patient genotype and coadministered drugs. Fig. 3. Scatter plot showing the relationship between log10 pharmacogeneticpredicted efavirenz mid-dose plasma concentrations (y-axis) and log10 observed concentration (x-axis) in 94 HIV-infected patients. Log10 efavirenz mid-dose plasma concentrations predictions for each subject were made based on their genotype carrier status (CYP2B6 c.516TT, CYP2A6*9 or *17 and/or UGT2B7*1a as indicated by arrows) using the pharmacogenetic algorithm derived by multiple linear regression analysis (model and associated goodness of fit statistics are shown at the top of graph). Veterinary pharmacogenomics Substantial differences in drug metabolism capacity also exist between different breeds of dog and between different species of animal. These phenomena are important to investigate for comparative purposes (especially humans versus laboratory animals) as well as for improving the health and welfare of our companion animals. Several projects are investigating why Greyhound dogs metabolize certain anesthetic agents more slowly compared with other dog breeds, and why cats have a poor capacity to metabolize certain drugs by glucuronidation. Fig. 4. Phylogram showing the established evolutionary relationships between species in the suborder Feloidea in relation to the appearance of disruptive mutations within the UGT1A6 gene. All Felidae share 5 mutations that likely originated between 11 and 35 million years ago resulting in UGT1A6 pseudogenization. MY - estimated times of divergence in millions of years ago.