* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download lecture13_2012
Survey
Document related concepts
Transcript
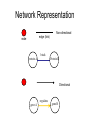
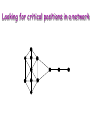
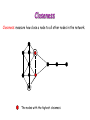
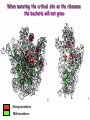
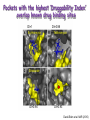
Biological Networks Can a biologist fix a radio? Lazebnik, Cancer Cell, 2002 Building models from parts lists Lazebnik, Cancer Cell, 2002 Building models from parts lists Computational tools are needed to distill pathways of interest from large molecular interaction databases Thinking computationally about biological process may lead to more accurate models, which in turn can be used to improve the design of algorithms Navlakha an Bar-Joseph 2011 The Protein-Protein Interaction Network in yeast Jeong et al. Nature 411, 41 - 42 (2001) Network Representation Non-directional edge (link) node binds protein A Protein B Directional regulates gene A gene B Different types of Biological Networks Nodes Edges Protein Interaction Transcriptional Proteins Transcription factor Target genes Physical Interaction Transcriptional Interaction Protein-Protein Protein-DNA A A B B Small-world Network Biological networks exhibit small-world network (SWN) characteristics (similar to social networks, internet etc) Every node can be reached from every other by a small number of steps SWN vs Random Networks Random Network Small World Network (SWN) SWN have a small number of highly connected nodes What can we learn from a network? What can we learn from Biological Networks Hubs are highly connected nodes • Hubs tend to be “older” proteins • Hubs are evolutionary conserved Are hubs functionally important ? Hubs are usually critical proteins for the species Lethal Slow-growth Non-lethal Unknown Jeong et al. Nature 411, 41 - 42 (2001) Networks can help to predict function Can the network help to predict function Begley TJ, Mol Cancer Res. 2002 •Systematic phenotyping of 1615 gene knockout strains in yeast •Evaluation of growth of each strain in the presence of MMS (and other DNA damaging agents) •Screening against a network of 12,232 protein interactions Mapping the phenotypic data to the network Begley TJ, Mol Cancer Res. 2002 Mapping the phenotypic data to the network Begley TJ, Mol Cancer Res. 2002 Networks can help to predict function Begley TJ, Mol Cancer Res. 2002. Case Study A network approach to predict new drug targets Aim :to identify critical positions on the ribosome which could be potential targets of new antibiotics Keats (1795-1821) Mozart (1756-1791) Kafka (1883-1924) Orwell (1903-1950) Schubert (1797-1828) Chopin (1810-1849) In our days… Infectious diseases are still number 1 cause of premature death (0-44 years of age) worldwide. Annually kill >13 million people (~33% of all deaths) The ribosome is a target for approximately half of antibiotics characterized to date Antibiotics targets of the large ribosomal subunit Looking at the ribosome as a network A1191 Looking at the ribosome as a network 1. Critical sites in the ribosome network may represent functional sites (not discovered before) 2. New functional sites may be good site for drug design Looking for critical positions in a network Looking for critical positions in a network Degree: the number of edges that a node has. The node with the highest degree in the graph (HUB) Looking for critical positions in a network Degree: the number of edges that a node has. The node with the highest degree in the graph (HUB) Closeness Closeness: measure how close a node to all other nodes in the network. The nodes with the highest closeness Betweenness Betweenness: quantify the number of all shortest paths that pass through a node. The node with the highest betweenness Looking for critical positions in a network The node with the highest degree The node with the highest betweenness The nodes with the highest closeness Looking at macromolecular structures as a network A1191 have the highest closeness, betwenness, and degree. A1191 How can the network approach help identify functional sites in the ribosome ? Characterize the whole ribosome as a network Calculate the network properties of each nucleotide ? Which (is there a?) property best characterizes the known function sites? When mutating the critical site on the ribosome the bacteria will not grow 2 Strong mutations Mild mutations 1 Critical site on the ribosome have unique network properties Strong mutations Mild mutations p~0 p~0 p=0.01 David-Eden et al, NAR (2008) ‘Druggability Index’ Based on the network property Bad site Good site David-Eden et al. NAR (2010) Pockets with the highest ‘Druggability Index’ overlap known drug binding sites DI=1 Erythromycin DI=0.98 Telithromycin Girodazole DI=0.94 DI=0.93 David-Eden et al. NAR (2010) Course Summary and How to start working on your project What did we learn • Pairwise alignment – Dynamic Programing Local and Global Alignments When? How ? Recommended Tools : for local alignment blast2seq last.ncbi.nlm.nih.gov/Blast.cgi?PAGE_TYPE=BlastSearch&PROG_DE F=blastn&BLAST_PROG_DEF=megaBlast&BLAST_SPEC=blast2seq For global best use MSA tools such as Clustal W2, Muscle (see next slide) What did we learn • Multiple alignments (MSA) When? How ? MSA are needed as an input for many different purposes: searching motifs, phylogenetic analysis, protein and RNA structure predictions, conservation of specific nts/residues Recommended Tools : Clustal W2 http://www.ebi.ac.uk/Tools/msa/clustalw2/ (best for DNA and RNA), MUSCLE http://www.drive5.com/muscle/ (best for proteins) Phylogeny.fr phylogenetic trees http://www.phylogeny.fr/ What did we learn • Search a sequence against a database When? How ? - BLAST :Remember different option for BLAST!!! (blastP blastN…. ), make sure to search the right database!!! DO NOT FORGET –You can change the scoring matrices, gap penalty etc - PSIBLAST Searching for remote homologies BLAST http://blast.ncbi.nlm.nih.gov/Blast.cgi What did we learn >Motif search When? How ? -Searching for overabundance of unknown regulatory motifs in a set of sequences ; e.g promoters of genes which have similar expression pattern (MEME) >Domain search Pfam (database to search for protein domains) Suggested Tools : MEME http://meme.nbcr.net/meme/ DRIMUST http://drimust.technion.ac.il/ PFAM http://pfam.sanger.ac.uk/ What did we learn • Protein Secondary Structure PredictionWhen? How ? – Helix/Beta/Coil – Most successful approaches rely on information from the environment and MSA - Predictions level around 80% Suggested tools GOR: Jpred: http://gor.bb.iastate.edu/ http://www.compbio.dundee.ac.uk/www-jpred/ What did we learn • Protein Tertiary Structure PredictionWhen? How ? – First we must look at sequence identity to a sequence with a known structure!! – Sequence homology based methodsHomology modeling – Structure homology based methods- Threading Remember : Low quality models can be miss leading !! Database and tools Protein Data Bank http://www.rcsb.org/pdb/home/home.do Suggested tool for molecular visualization http://www.pymol.org/ Good tool for homology modeling http://swissmodel.expasy.org/ What did we learn • RNA Structure and Function PredictionWhen? How ? – MFE based methods– good for local interactions, several predictions of low energy structures – Adding information from MSA can help but usually not available – RNA families are characterized by their structure (Rfam). Suggested tools: RNAfold http://rna.tbi.univie.ac.at/cgi-bin/RNAfold.cgi RFAM http://rfam.sanger.ac.uk/ What did we learn • Gene expression When? How ? > Unsupervised methodsDifferent clustering methods : K-means, Hierarchical Clustering > Supervised methods-such as SVM – GO annotation (analysis of gene clusters..) Selected databases and tools GEO http://www.ncbi.nlm.nih.gov/geo/ EPclust http://www.bioinf.ebc.ee/EP/EP/EPCLUST/ David http://david.abcc.ncifcrf.gov/ Most useful databases Genomic database The human genome browser http://genome.ucsc.edu/ Protein database Uniprot http://www.uniprot.org/ Structure database PDB (RCSB) http://www.rcsb.org Gene expression database GEO http://www.ncbi.nlm.nih.gov/geo/ So How do we start … Now that you have selected a project you should carefully plan your next steps: A. Make sure you understand the problem and read the necessary background to proceed B. formulate your working plan, step by step C. After you have a plan, start from extracting the necessary data and decide on the relevant tools to use at the first step. When running a tool make sure to summarize the results and extract the relevant information you need to answer your question, it is recommended to save the raw data for your records , don't present raw data in your final project. Your initial results should guide you towards your next steps. D. When you feel you explored all tools you can apply to answer your question you should summarize and get to conclusions. Remember NO is also an answer as long as you are sure it is NO. Also remember this is a course project not only a HW exercise. . Example • Amyloids are proteins which tend to aggregate in solution. Abnormal accumulation of amyloid in organs is assumed to play a role in various neurodegenerative diseases. Question : can we predict whether a protein X is an amyolid ? Preparing a poster Prepare in PPT poster size 90-120 cm Title of the project Names and affiliation of the students presenting The poster should include 5 sections : Background should include description of your question (can add figure) Goal and Research Plan: Describe the main objective and the research plan Results (main section) : Present your results in 3-4 figures, describe each figure (figure legends) and give a title to each result Conclusions : summarized in points the conclusions of your project References : List the references of paper/databases/tools used for your project Key date reminder 22.1 Submission project overview (one page) -Title -Main question -Major Tools you are planning to use to answer the questions Final week – meetings on projects 14.3 Poster submission 20.3 Poster presentation