* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Enzymes: Biological Catalysts
Survey
Document related concepts
Transcript
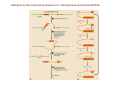
Enzymes: Biological Catalysts 10/11/07 The Role of Enzymes • A catalyst increases the rate or velocity of a chemical reaction without itself being changed in the overall process. Most biological catalysts are proteins called enzymes. The substance acted on by an enzyme is called a substrate. Enzymes speed up reactions by many orders of magnitude. For example, the enzyme catalase speeds up the conversion of hydrogen peroxide to water and oxygen by a factor of a billion. • True catalysts, such as enzymes, participate in the reaction, but are unchanged by it. Therefore, they can continue to catalyze subsequent reactions. • Catalysts change the rates of reactions, but do not affect the equilibrium of a reaction. i.e., you cannot make more product from an enzyme-catalyzed reaction than you can from the same reaction without it. The enzyme simply helps to reach the equilibrium state faster than if it were not present. Reaction Order • • • First order reactions - For the irreversible reaction, A Æ B, the reaction rate (V) is given by V = d[B]/dT (for the rate of appearance of the product, B or by V = -d[A]/dT (for the rate of disappearance of the substrate, A). These equations are equally valid for this reaction, so V = d[B]/dT = -d[A]/dT = k1[A], where k1 is called the rate constant, (seconds)-1. The first-order reaction called, is due to the first power of the reactant concentration. If k1 is large, the reaction is fast and if k1 is small, the reaction is slow. Integrating the above equation yields [A]/[A]0 = e-(k t), where [A]0 is the starting concentration of A when t = 0. A plot of this equation (Figure a) shows that the concentration of A decreases exponentially with time. The amount of time it takes for half of A to be lost is called the half-life and is given by t1/2. The half-life is inversely proportional to k1. A plot of ln[A] vs t, as shown in Figure b, will always yield a straight line with slope = -k1 if the reaction is first order. If one plots the initial rate of the reaction versus varying starting concentrations ([A]0), a straight line plot with slope of k1 will be produced for a first order reaction. The most common example of a first order reaction is the decay of radioactive elements. 1 • • Determining the order and rate constant of an irreversible firstorder reaction. The equilibrium does not lie far to one side and, as product accumulates, the reverse reaction becomes important. So, for the reaction A kk1 B, a forward -1 reaction constant, k1, can be used to define the reaction moving rightward and a reverse reaction constant, k -1 can be used to define the reaction moving leftward. Now the molecule A is being consumed in the reaction to the right and formed by the reaction to the left, so the corresponding rate equation is -V = d[A]/dt = -k1[A] + k-1[B] Such a reaction approaches a state of equilibrium, at which point the rates of the forward and reverse reactions become equal. At the same time, the overall rate becomes zero, so 0 = -k1[A]eq + k-1[B]eq or [B]eq/[A]eq = k1/k-1 = K, where K is the equilibrium constant. For a reversible reaction that is first-order in both directions, the equilibrium constant is always the ratio of the forward and reverse rate constants. Reaction Order • Second-Order Reactions - A reaction of this type typically occurs when two molecules come together to form products. A simple example is k2 2A Æ A2 • with a rate constant given by k2. The rate of such a reaction is proportional to the second power of the concentration of the reactant. Therefore, V = -d[A]/dt = -k2[A]2 • Here k2 is the second-order rate constant. It has dimensions of (mol/L)-1s-1 or M-1s-1 Transition States and Reaction Rates • The below Figure depicts a plot of the free energy of the system versus the reaction coordinate for a chemical reaction. This simplistic view illustrates that the standard state free energy of the products is lower than that of the reactants. It shows little of what happens in the transition from reactant to product. • Reactions do not occur all at once. A molecule in a first-order reaction must only occasionally reach an energy state in which the process can occurotherwise all molecules would react at once. In reality, only a certain fraction of the molecules which are sufficiently energetic can undergo reactions. The same is true in second order reactions Reactions have free energy "barriers" to them. An activated or "transition state" for a molecule occurs when it has reached an energy that is sufficient to react. Figure b more accurately depicts the free energy barrier for a reaction. Figure c shows the same conversion for an actual molecular alteration-a boat Æ chair conversion of a pyranose ring. Here the transition state is drawn as a half-boat/half-chair structure. As shown in Figures b and c, the transition state (symbolized by ‡ ) represents an intermediate molecular state having an increase in free energy of ΔG1o‡ above that of the initial substrate. If the barrier to reaction is high, only a small fraction of the molecules will have enough energy to react. • If let [A]‡ represent the concentration of molecules having the activation energy, then we can write the equilibrium constant K as [A]‡ /[A]. Substituting into equation 3.23, we obtain [A] = [A]e- /RT Because only molecules at the transition state can proceed to react (in either direction), the rate constant can be then expressed as proportional to the population of the transition state: , where Q represents the frequency of forming the product (right side of reaction equation). Substituting ΔG1o‡ = ΔHo‡ - TΔSo‡ , we obtain which is known at the Arrhenius equation. Q' is a constant equal to Q’ = Qe(ΔSo‡/R) so we take the natural logarithms of both sides and simplify to obtain Since we expect ΔH。‡ to be positive, reactions should go faster at higher temperatures. From equation 11.11, a graph of ln k versus 1/T should be a straight line and its slope should give Δ H。‡ . An example (the reaction of L-malate to yield fumarate and water) is shown in Figure below. The activation energy opposes the reaction in both directions, with ΔG1o‡ and ΔG-1o‡ representing the transition state free energy barrier in the forward and reverse directions, respectively. Incorporating this and K = k1/k-1, - = , Thus, the equilibrium constant K tells nothing about the rates of process. K depends only on the free energy difference between final and initial states and carries no information about the height of the barrier between these states. Summary of important points: 1. Reactions should go faster at higher temperatures. 2. The activation energy opposes the reaction in both directions. 3. K reveals nothing about the rates of process-it depends only on the free energy difference between final and initial states and carries no information about the height of the barrier between these states. How Catalysts Work • • A catalyst works simply by lowering the energy barrier of a reaction. By doing so, the catalyst increases the fraction of molecules that have enough energy to attain the transition state, thus making the reaction go faster in both directions. The position of the equilibrium (the amount of product versus reactant) is unchanged by a catalyst. Even though k1 and k-1 many be greatly changed from their values in the absence of a catalyst, each one changes by the same factor and the equilibrium constant, K, is unchanged, because K = k1/k-1. Catalysts lower the energy barrier in two ways: 1. The catalyst binds a substrate in an intermediate conformation that resembles the transition state, but has a lower energy. This may lead to multiple intermediate states that bypass the transition state. An intermediate state is a metastable state of a molecule. 2. In a non-catalyzed reaction the entropy may be highly negative due to the highly specific orientation required in order for a reaction to occur. (Note that a more negative entropy contributes to a more positive free energy of transition.) Catalysts can lower the negative entropy by binding reacting molecules only in the proper mutual orientation, thus increasing their reactivity. The Induced Fit Model • • The induced fit model of enzyme action is a modification of the lock-andkey model originally proposed by Emil Fischer in 1894. The lock-and-key model proposes that an enzyme/substrate pair is like a lock and key. Though it explains the specificity of enzyme/substrate pairs, it does little to explain catalysis, because a lock does not change a key the way an enzyme changes a substrate. In 1958, Daniel Koshland proposed the induced fit model to explain enzymatic catalysis. The model proposes that distortion of the enzyme and the substrate is an important event in catalysis. Figure below shows x-ray diffraction studies of the enzyme hexokinase both without (a) and with (b) glucose bound. Note that binding of glucose causes two domains of the enzyme to fold toward each other. Enzymes do more than simply bind and position substrates, however. Enzymes 1. Bind substrate(s); 2. Lower the energy of the transition state; and 3. Directly promote the catalytic event. The latter may occur as a result of specific amino acid side chains that physically interact with the substrate and end up promoting the reaction. When the catalytic process is complete, the enzyme must be able to release the product or products and return to its original state for another round of catalysis. For an enzyme (E) that catalyzes the conversion of a single substrate (S) into a single product (P), the simplest way to write the overall reaction is in two steps: S + E <=> ES ES Æ E + P Here, ES represents the enzyme-substrate complex. For simplicity, the first reaction is shown as reversible, while the second reaction is irreversible. For specific examples, see the triose phosphate isomerase and serine proteases below. Triose Phosphate Isomerase Catalysis • • • Triose phosphate isomerase catalyzes the following reaction: Glyceraldehyde-3-Phosphate (G3P) <=> cis-enediol intermediate <=> Dihydroxyacetone Phosphate (DHAP) As the reaction is written, consider G3P as the substrate and DHAP as the product. The enediol intermediate is unstable and normally has a much more positive free energy than either G3P or DHAP. The active enzyme is a dimer of two identical subunits. Each has the configuration shown in Figure a. The active site (the place on the enzyme where catalysis occurs) can accommodate either G3P or DHAP. At the active site, a glutamate residue (Glu 165) and a histidine (His 95) are essential for function of the enzyme. After binding, an enzyme "lid" closes down on the substrate to provide a cage that protects the enediol intermediate. In the absence of the lid, the enediol intermediate is lost and catalytic efficiency decreases by a factor of 100,000. Conversion of Glu 165 to Asp retains the negative charge, but reduces efficiency by a factor of 1000. The reaction can be broken down into the following steps: E + G3P <=> E-G3P (Binding of G3P) E-G3P <=> E-ed (Conversion to enediol) E-ed <=> E-DHAP (Conversion to DHAP) E-DHAP <=> E + DHAP (Release of DHAP) Like other enzymes, triose phosphate isomerase lowers the energy barriers of the transition states. Triose phosphate isomerase is an extremely efficient enzyme. Serine Protease Catalysis • Serine proteases are enzymes that catalyze the hydrolysis of peptide bonds. In each case, the enzymes have a serine residue that plays a critical role in the catalysis. The enzymes cuts preferentially in distinct sites. The active site regions of all of the serine proteases have a number of common factors. For example, an aspartate residue, a histidine residue, and a serine residue are always clustered about the active site depression. Such a structure for chymotrypsin is shown in below Figure. Also, a "pocket" is always located close to the active site serine. Active site pockets of two serine proteases. •The shape and charge of the "pocket," however, vary between different serine proteases. Thus, it is the nature of the pocket that gives a serine protease its specificity. For example, in chymotrypsin, the pocket is wide and lined with hydrophobic residues to accommodate a hydrophobic side chain, such as phenylalanine. The catalytic mechanism of chymotrypsin, a serine protease is shown in nest slide. These steps include the following: 1. Polypeptide substrate binding. 2. Proton transfer from Ser to His. The substrate forms a tetrahedral transition state with the enzyme. 3. Proton transfer to the C-terminal fragment, which is released by cleavage of the C-N bond. The N-terminal peptide is bound through acyl linkage to serine. 4. A water molecule binds to the enzyme in place of the departed polypeptide. 5. The water molecule transfers its proton to His 57. Again, a tetrahedral transition state is formed. 6. The second peptide fragment is released. The acyl bond is cleaved, the proton is tranferred from His back to Ser, and the enzyme returns to its initial state. A key to the mechanism of serine protease catalysis lies in the stability of the two tetrahedral intermediate states, which are very similar to the essential transition states. They appear to be stabilized by hydrogen bonds from backbone amino protons from residues Ser 195 and Gly 193 to one of the oxygens in the tetrahedral complex (the carbonyl oxygen of the substrate). The hydrogen bonding can occur only with formation of the tetrahedral state and thus stabilizes the intermediates. Both triose phosphate isomerase and serine proteases have a histidine and an acidic residue in their active site. Histidine is very common in active sites, because it readily accepts or donates protons at physiological pH. Catalysis of peptide bond hydrolysis by chymotrypsin. Michaelis-Menten Kinetics • Equation above represents the minimal equation needed to describe a simple one-substrate, one-product reaction catalyzed by an enzyme. • This assumes the reverse reaction between E and P is negligible and is a simple, first-order reaction whose rate is determined by [ES] and the value of k2. The reaction rate can be written as V = k2[ES] • [ES] is not a measurable concentration. Measurable items are the [S] and [E]t , which is the sum of the free enzyme and complexed enzyme. i.e. [E]t = [E] + [ES] • It is incorrect to assume that E and S are in equilibrium with ES, because some ES is continually being drained off to make P. • Briggs and Haldane assumes that when the reaction is started by mixing enzymes and substrates, the ES concentration builds up at first, but quickly reaches a steady state, in which it remains almost constant. The steady state will persist until almost all of the substrate has been consumed. If one measures rates only after the steady state has been established and before [ES] has changed much, reaction velocity can be calculated by assuming steady state conditions. The steady state in enzyme kinetics. In the steady state, rates of formation and breakdown of ES are equal, Rearranging 11.17 gives equation 11.18, Combining the three rate constants into one, KM, yields KM = (k-1 + k2)/k1 Equation 11.18 becomes: KM[ES] = [E][S] Because [E] = [E]t - [ES], KM[ES] = [E]t[S] - [ES][S] Solving for [ES], [ES] = [E]t[S]/(KM+[S]) Substituting this into the earlier velocity equation, V = k2[E]t[S]/(KM+[S]) This is the Michaelis-Menten equation, and KM is called the Michaelis constant. KM has units of concentration and, because it is a ratio of the rate constants of a reaction, KM is characteristic of the reaction. a plot of velocity (V) versus substrate concentration ([S]) Note that at high substrate concentrations ([S] >> KM), the velocity approaches a maximum (called Vmax). Note also that the substrate concentration where V = Vmax/2 corresponds to the KM. At Vmax, [S] >> KM, so [S] + KM ≒ [S]. Thus, the velocity equation simplifies to Vmax = k2[E]t Substituting this back into the velocity equation yields V = Vmax[S]/(KM + [S]) k1 k2 E+S ES E+P k-1 Vmax = k2[E]t V = k2[ES] V = Vmax[S]/(KM + [S]) [E]t = [E] + [ES] Ks = k-1/k1 = [E][S]/[ES] k1[E][S] = k-1[ES] + k2 [ES] [ES] = (k1/(k-1 + k2)) [E][S] KM = (k-1 + k2)/k1 KM[ES] = [E][S] KM[ES] = [E]t[S] – [ES][S] [ES](KM +[S]) = [E]t[S] [ES] = [E]t[S]/(KM +[S]) V = k2[E]t[S]/(KM + [S]) (d[ES]/dt = k1[E][S]-k-1[ES]-k2[ES]) (In a steady state, d[ES]/dt = 0) For a multistep reaction, the Michaelis-Menten equation must be modified. Consider the reaction For this reaction, the k2 in the Michaelis-Menten equation must be replaced by a more general constant, called kcat. Here, V = kcat[E]t[S]/( KM + [S]) kcat incorporates the rate constants for all the reactions between ES and E + P. For the two-step reaction above, kcat = k2. For more complex reactions, kcat depends on which steps in the process are rate-limiting. KM, kCAT, and kCAT/KM • For reactions obeying Michaelis-Menten kinetics, KM is a measure of the substrate concentration required for effective catalysis to occur (the affinity of the enzyme for substrate ). i.e., an enzyme with a high KM requires a higher substrate concentration to achieve a given reaction velocity than an enzyme with a low KM. Table below lists values of KM, kcat, and kcat/KM for selected enzymes. kcat Æ V = kcat[E]t[S] / ( KM + [S]) (11.27) kcat gives a direct measure of the catalytic production of product under optimum conditions (saturated enzyme). The units of kcat are second s-1. The reciprocal of kcat can be thought of as the time required by an enzyme molecule to "turn over" one substrate molecule. Alternatively, kcat measures the number of substrate molecules turned over per enzyme molecule per second. Thus, kcat is sometimes called the turnover number. kcat/KM Æ a measure of enzyme efficiency. Either a large value of kcat (rapid turnover) or a small value of KM (high affinity for substrate) makes kcat/KM large. When [S] << KM (dilute solution), V≒ (kcat/KM)[E][S] Here, kcat/KM behaves as a second-order rate constant for the reaction between substrate and free enzyme. This ratio is important, for it shows what the enzyme and substrate can accomplish when abundant enzyme sites are available, and it allows direct comparison of the effectiveness of an enzyme toward different substrates. When an enzyme has a choice of two substrates, A or B, present at equal, dilute concentrations, Table shows that when an enzyme has different substrates on which it can work. Note that for chymotrypsin the kcat/KM ratio varies 1-million fold. As a second-order rate constant, kcat/KM has a maximum possible value, which is determined by the frequency with which enzyme and substrate molecules can collide. A reaction which attains such a velocity is said to be "diffusion-limited" because every encounter leads to reaction. If every collision results in formation of an enzyme-substrate complex, diffusion theory predicts that kcat/KM will attain a value of about 108 to 109 (mol/L)-1s-1. The enzymes carbonic anhydrase, fumarase, and triose phosphate isomerase actually approach this limit. Analysis of Kinetic Data • • To test whether an enzyme-catalyzed reaction follows the Michaelis-Menten law, the initial rates of a series of reactions are measured (all at the same enzyme concentration, but at different substrate concentrations). Because the initial [S] is known precisely, and the change in [S] versus t is almost linear in the initial stages, accurate data for V as a function of [S] can be obtained. A Lineweaver-Burk plot is obtained by inverting the Michaelis-Menten equation to obtain the following: A Lineweaver-Burk plot. For a reaction obeying Michaelis-Menten kinetics, plotting 1/V versus 1/[S] should yield a straight line. Vmax and KM can be obtained from such a plot. An Eadie-Hofstee plot. A disadvantage of the Lineweaver-Burk plot is the long extrapolation necessary to determine KM. One way around this problem is to graph V versus V/[S] (called an Eadie-Hofstee plot). The result also yields a straight line for reactions obeying Michaelis-Menten kinetics. Here, the slope of the line is equal to -KM. Multisubstrate Reactions • Proteolysis involves two substrates (the polypeptide and water) and two products (the two fragments of the cleaved polypeptide chain). • When an enzyme binds two or more substrates and releases multiple products, the order of the steps becomes an important feature of the enzyme mechanism. Several classes of mechanisms include the following: 1. Random substrate binding - In this mechanism, either substrate can be bound first, though in many cases one substrate will be favored for initial binding, and its binding may promote the binding of the other. The general pathway is The phosphorylation of glucose by ATP, catalyzed by hexokinase, appears to follow this mechanism, with some tendency for glucose to bind first. 2. Ordered substrate binding - This mechanism occurs when one substrate must bind before a second one can bind significantly. This mechanism is Ordered substrate binding is often observed in oxidations of substrates by the coenzyme nicotinamide adenine dinucleotide (NAD+). 3. The "ping-pong" mechanism - This occurs when a catalytic sequence of events occurs, such as one substrate is bound, one product is released, a second substrate is bound, and a second product is released. This is shown as where E* is a modified form of the enzyme, often carrying a fragment of the first substrate, S1. A good example is the cleavage of a polypeptide chain by a serine protease, such as trypsin or chymotrypsin. The polypeptide is described here as S = A.B, where A and B designate the C-terminal and N-terminal portions of the chain from the point of cleavage: Here E*.B and E*.B.H2O indicate covalent intermediates, as in Figure below. Kinetics of a Complex Reaction - For the cleavage of a substrate by a serine protease, such as chymotrypsin, the step E*.B + H2O Æ E*.B.H2O cannot be analyzed. Since the concentration of water is essentially fixed in aqueous solution and is not a variable, the reaction can be written as Steady state measurements in this case will be insufficient. The steady state velocity is given by The enzyme obeys Michaelis-Menten kinetics, but kcat, KM, and Ks are defined as kcat = k2k3/(k2 + k3) KM = Ksk3/(k2 + k3) Ks = k-1/k1 Thus, the Michaelis-Menten equation describes the velocity correctly, but the values of kcat and KM depend on the reaction mechanism. To obtain the individual rate constants in such a case, measurements outside the steady state range must be employed. The kinetics of the hydrolysis of esters by chymotrypsin (the enzyme also works on esters) reveals a rapid concentration increase for a few minutes until about one molecule has been produced per enzyme molecule. Steady state production begins after that point. The initial burst is called pre-steady state production. For ester hydrolysis, k3 is much smaller than k2. Thus, the acyl intermediate forms quickly on each enzyme molecule, with accompanying release of product A. After this period, however, more A can be formed only after each acyl intermediate breaks down and the enzyme becomes available again. The dissociation of the acyl intermediate is the rate-limiting step. Faster measurement techniques, such as stopped-flow methods, allow measurement of the rate of formation of the ES complex. Measurements of the decay of the acyl intermediates after substrate is exhausted provide k3. Combinations of these methods can be used to obtain all of the constants in equation 11.34. Example rate constants for hydrolysis of two N-acyl amino acid esters by chymotrypsin are given in Table. Enzyme Inhibition • Many different kinds of molecules inhibit enzymes and the inhibition may be reversible or irreversible. Reversible inhibition involves noncovalent binding of the inhibitor and can always be reversed by removal of the inhibitor. Irreversible inhibition involves a covalent binding of a molecule to the enzyme, which incapacitates it. Reversible Inhibition • Competitive inhibition - the inhibiting compound so closely resembles the substrate for the enzyme that it accepts the molecule to the substrate binding site. However, once bound, the inhibitor cannot be acted on and thus prevents the enzyme from catalyzing the intended reaction. This reaction scheme is depicted as I stands for the inhibitory compound and KI is the dissociation constant for inhibitor binding -KI = [E][I]/[EI]. Now, [E]t = [E] + [ES] + [EI], where [EI] is the concentration of inhibitor-enzyme complex. Then, the velocity is where is the apparent KM given by = KM(1 + [I]/KI) Increasing [I] causes an apparent increase in the KM. The Vmax is unchanged, because as [S] increases relative to a fixed [I], the substrate molecules outcompete the inhibitor for the enzyme's active site. The effect of a competitive inhibitor on a graph of V versus [S] is shown in Figure a. The system still obeys an equation of the Michaelis-Menten form at a given [I], so the Lineweaver-Burk and Eadie-Hofstee plots are linear, but the KM is altered (Figure b). By plotting the apparent KM as a function of [I] (Figure 1c), one can obtain both KM and KI. A variant of competitive inhibition is nonproductive binding. This occurs when a substrate molecule can fit into the enzyme's second binding site in such a way that the normal catalytic event cannot occur. This scheme is as follows: Here, ES' is the enzyme-substrate complex that cannot lead to product. In this situation, both KM and kcat are modified. Noncompetitive inhibition - This occurs when a molecule or an ion binds to a site on the enzyme other than the active site and modifies kcat (Figure 11.22). Such a compound need not resemble the substrate at all. In fact, it only needs to have a strong affinity for the second binding site. Assuming the inhibitor has equal affinity for E and ES, the scheme is Noncompetitive inhibition. Mathematical analysis yields equation 11.38. the KM is unaffected, but the apparent kcat is now given by kappcat=kcat/([1+[I]/KI]). The apparent kcat decreases with increasing [I]. Vmax is also changed (Figure 11.23a): Vappmax=kcat(apparent)[E]t = kcat[E]t/(1 + [I]/KI) The effect of noncompetitive binding on a Lineweaver-Burk plot is shown in Figure 11.23b. Both the true kcat and KI are determined by graphing 1/Vappmax versus [I] (Figure 11.23c). Effects of noncompetitive inhibition on enzyme kinetics. The situation is usually more complex than shown here. For example, the complex ESI may also be able to undergo the catalytic process slowly, or the binding of inhibitor may modify both kcat and KM. The latter case is called mixed inhibition. Irreversible Inhibition • • Irreversible inhibition occurs when substances combine covalently with enzymes so as to inactivate them irreversibly. Almost all irreversible enzyme inhibitors are toxic substances, either natural or synthetic. Some, such as cyanide and penicillin, are shown in next slide. Figure in next slide depicts the action of the competitive irreversible inhibitor, diisopropyl fluorophosphate (DFP), which reacts with serine groups on a protein to form a covalent adduct. Irreversible inhibitors that strongly resemble the substrate rather than its transition state may be extremely selective. An example is TPCK in next slide, which is an excellent inhibitor for chymotrypsin. When selective irreversible inhibitors are used to label active site residues of an enzyme to aid in their identification, they are called affinity labels. A suicide inhibitor, on the other hand, is an affinity label that is unreactive until it is acted upon by the enzyme, at which point it binds. Irreversible inhibition by adduct formation. Function of Coenzymes A protein may require the help of some other small molecule or ion to carry out the reaction. Molecules bound to enzymes for this purpose are called coenzymes. The water soluble vitamin B complexes are metabolic precursors of a number of coenzymes. Table lists several important coenzymes together with their related vitamins. NAD+ - Nicotinamide adenine dinucleotide (NAD+) is derived from the vitamin niacin. The nicotinamide portion of the molecule is capable of being reduced and can thus serve as an oxidizing agent, where 'R' stands for the remainder of the molecule. NAD+/NADH behave both like a second substrate in a reaction (because each is converted to the other by the enzyme) and like a coenzyme (because they are recycled over and over). They are generally classified as coenzymes Metal Ions in Coenzymes - Many enzymes contain metal ions, usually held by coordinate covalent bonds from amino acid side chains, but sometimes bound by a prosthetic group like heme. Such enzymes are called metalloenzymes. Figure below shows the active site of the protease carboxypeptidase A, which contains a zinc ion. Classification of Protein Enzymes 1. 2. 3. 4. 5. 6. The Enzyme Commission (EC) of the International Union of Biochemistry and Molecular Biology (IUBMB) devised a naming and numbering system. Eenzymes are divided into six major classes, with sub-groups and subsubgroups to define their functions more precisely. The major classes are as follows: Oxidoreductases catalyze oxidation - reduction reactions. Transferases catalyze transfer of functional groups from one molecule to another. Hydrolases catalyze hydrolytic cleavage. Lyases catalyze removal of a group from or addition of a group to a double bond, or other cleavages involving electron rearrangement. Isomerases catalyze intramolecular rearrangement. Ligases catalyze reactions in which two molecules are joined. The EC of the IUBMB has given each enzyme a number with four parts, such as EC 3.4.21.5. The first three numbers define major class, subclass, and sub-subclass, respectively. The last is a serial number in the subsubclass, indicating the order in which each enzyme is added to the list, which is continually growing. For example, triose phosphate isomerase is listed as EC 5.3.1.1. Thus, it is an isomerase and in the third subclass (enzymes that involve an oxidation in one part of the substrate molecule and reduction in another). It is in the first sub-subclass (those that interconvert aldoses and ketoses) and is the first entry (of 19 so far) in this sub-subclass. Molecular Engineering Molecular engineering is a term used to describe the design of enzymes using modern molecular biological techniques to alter their catalytic action. Examples include the following: • Site-directed mutagenesis - The DNA coding sequence for an enzyme is altered to change one or more amino acids in an enzyme when the mutated DNA is expressed in an organism. • Hybrid enzymes - Molecular techniques are used to put together two different biomolecules to make a fusion molecule with new, useful properties. Figure in next slide depicts a hybrid enzyme made in this fashion. In this case, an oligonucleotide of a defined sequence has been grafted onto the enzyme staphylococcal nuclease. The specific sequence in the hybrid enzyme allows it to bind to a specific complementary nucleic acid sequence (specified by the bound oligonucleotide) and cut specifically at that point. The native, unaltered enzyme has no such specificity. • Catalytic antibodies - Antibodies have a very specific binding site to the transition state of an enzymatic reaction. The resulting molecules, called abzymes, act like antibodies. In some cases, abzymes can speed up reaction rates as much as 107-fold over the uncatalyzed reaction. The stereospecificity of enzymes (including abzymes) may provide a tremendous aid to the synthesis of stereospecific compounds in organic chemistry. A hybrid enzyme. Ribozymes • Some RNA molecules, called ribozymes are capable of catalyzing chemical reactions too. Figure 11.29 shows the site of action of the RNA-protein complex called ribonuclease P. The RNA portion of the complex can, by itself, catalyze the hydrolysis of the specific bond indicated by the red wedge in the figure. Tom Cech identified an interesting protein-independent self-splicing agent from the preribosomal RNA of the protist, Tetrahymena. In this reaction, the rRNA itself catalyzes removal of an RNA intron from itself. The RNA molecule involved in the catalysis is altered, so it is not technically considered a catalyst, but the sequence which is removed (called L-19 IVS) does have true catalytic activity. It can either lengthen or shorten small oligonucleotides, in the manner shown in next slide. Catalysis by the intervening sequence in Tetrahymena preribosomal RNA. Regulation of Enzyme Activity • Coordinating and regulating enzymatic activities is essential for efficient functioning of cells. Several control mechanisms that do not involve covalent modification of the enzymes are possible: 1. Substrate level control - High levels of the product of a reaction inhibit the ability of the small amounts of substrate present to react. An example is the first step in glycolysis, catalyzed by hexokinase. It is inhibited by the product of its action, glucose-6-phosphate. If glycolysis is blocked for any reason, glucose-6-phosphate accumulates. 2. Feedback control - The product of a series of reactions inhibits the action of an earlier step in the process (usually the first step). Feedforward regulation occurs when a molecule in an assembly line reaction activates an enzyme ahead of it in the pathway. 3. Allosteric enzymes - These enzymes are invariably multisubunit proteins, with multiple active sites. They exhibit cooperativity in substrate binding (homoallostery) and regulation of their activity by other, effector molecules (heteroallostery). Homoallostery - The effects of cooperative substrate binding on enzyme kinetics are shown in Figure. Binding of one substrate favors binding of additional substrates. Cooperative binding favors reduction of KM for the binding of substrates after the initial one. The Figure shows the effect of extreme homoallostery. At concentrations of S below a critical point, [S]c, the enzyme is almost inactive, but then changes activity rapidly with concentrations of S greater than [S]c. Heteroallostery - This type of allosteric control involves heteroallosteric effectors which may be either inhibitors or activators of binding. If an enzyme can exist in two conformational states, T and R, that differ dramatically in the strength with which substrate is bound or which differ significantly in the catalytic rate, then the kinetics of the enzyme can be controlled by any other substance that, in binding to the protein, shifts the T<==>R equilibrium. Allosteric inhibitors shift the equilibrium toward T and activators shift it toward the R state. The Figure illustrates a V-vs-[S] curve. Note that shifts toward the R state (activators) increase the velocity for a given substrate concentration, whereas shifts toward the T state have the opposite effect. Aspartate Carbamoyltransferase • Aspartate carbamoyltransferase (also known as aspartate transcarbamoylase or ATCase) is known for its allosteric regulation: Aspartate + Carbamoyl Phosphate <==> Carbamoyl Aspartate ATCase is at the crossroads of biosynthetic pathways that lead to proteins or pyrimidines. ATCase catalyzes the joining of aspartate and carbamoyl phosphate to form N-carbamoyl-L-aspartate, the first metabolite committed to the synthesis of pyrimidines. The enzyme is sensitive to feedback inhibition by cytidine triphosphate (CTP), and is activated by ATP. Control points in pyrimidine synthesis. Regulation of aspartate carbamoyltransferase by ATP and CTP. Allosteric regulation of ATCase involves changes in the quaternary structure of the molecule. A major rearrangement of subunit positions occurs in the TÆ R transition. Quaternary structure of aspartate carbamoyltransferase (ATCase). ATCase is a multisubunit protein consisting of 6 catalytic subunits and 6 regulatory subunits. (The quarternary structure of ATCase enzyme is shown). The detailed structure of one catalytic subunit (green) and adjacent regulatory subunit (yellow) of ATCase. Sometimes different organisms regulate similar pathways in different ways. For example, ATCase is the major control point in the pyrimidine pathway synthesis in bacteria, whereas eukaryotes regulate the synthesis of carbamoyl phosphate. In mammals, the carbamoyl phosphate synthetase II is inhibited by UDP, UTP, CTP, dUDP, and UDP-glucose. Covalent Modifications to Regulate Enzyme Activity • • • Covalent modification activates some enzymes and inactivates others. i.e. some enzymes are wholly inactive until they are covalently modified and then begin to function. In other cases, covalent modification acts in the opposite direction, to inactivate otherwise active enzymes. Some such modifications can be reversed; others cannot. One of the most widespread modifications is phosphorylation or dephosphorylation of various amino acid side chains (e.g., serine, threonine, tyrosine, and histidine). These kinds of modification are most often a part of complex regulatory pathways, frequently under hormonal control. Proteolytic cleavage, eg. pancreatic proteases (such as trypsin, chymotrypsin, elastase, and carboxypeptidase): They are made as longer, catalytically inactive molecules called zymogens (trypsinogen, chymotrypsinogen, proelastase, and procarboxypeptidase, respectively). The zymogens must be cleaved proteolytically in the intestine to yield the active enzymes. If a small amount of protease becomes active in the pancreas, it can have painful or fatal consequences (i.e., acute pancreatitis). The pancreas protects itself from active protease action by synthesis of a protein called the secretory pancreatic trypsin inhibitor. The binding between trypsin and its inhibitor is one of the strongest noncovalent interactions known in biochemistry. The intestinal tissue is protected from damage by proteases by its glycosylated surface. Zymogen activation by proteolytic cleavage. The first step is activation of trypsin in the duodenum. A hexapeptide is removed from the N-terminal end of trypsinogen by enteropeptidase, a protease secreted by duodenal cells. This yields active trypsin, which then activates the other zymogens by specific proteolytic cleavages. Trypsin will also activate other trypsinogen molecules in an autocatalytic process. Activation of a few trypsinogens ultimately leads to activation of many trypsins. Activation of chymotrypsinogen is shown in next slide. First, trypsin cleaves the bond between arginine 15 and isoleucine 16. Notice that the N-terminal peptide remains attached to the rest of the molecule due to the disulfide bond between residues 1 and 122. The enzyme is activated by the cleavage due to changes in the conformation of the molecule. These include: 1. Creation of a new, positively charged N-terminal residue at Ile 16; 2. Salt bridge formation between Ile 16 and Asp 194 (next to the active site); 3. Movement of active site residues so that the amino groups of residues 193 and 195 are properly positioned to hydrogen-bond to the substrate oxyanion in the tetrahedral transition state. 4. Autocatalytic cleavages to remove residues 14-15 and 147-148 from the molecule produce the final, active form of chymotrypsinogen, called -chymotrypsin. Activation of chymotrypsinogen. Blood Clotting • Blood clots are composed of striated fibers of a protein called fibrin. Fibrin fibers are derived from a zymogen precursor, called fibrinogen, by proteolytic cleavages that release fibrinopeptides A and B from the molecule. Removal of these small fibrinopeptides exposes sites in the fibrin molecules that allows them to stick together. Covalent cross-links between glutamine and lysine residues also form to help stabilize the structure. Thus, activation of zymogens is a key aspect to clotting of blood in vertebrates. 1. 2. 3. 4. 5. 6. 7. 8. production of fibrin is the product of action of a cascade of proteases as follows: In damaged tissue, the proteins kininogen and kallikrein activate factor XII (part of the intrinsic pathway). Factor XII activates factor XI (part of the intrinsic pathway). Alternatively, damage to blood vessels leads to release of tissue factor and activation of factor VII (start of the extrinsic pathway). The extrinsic and intrinsic pathways merge with activation of factor X. Activation of factor X by factor IX of the intrinsic pathway requires factor VIII (antihemophilic factor). Factor X activates prothrombin to thrombin. Thrombin removes small fibrinopeptides from fibrinogen to form fibrin. Note that the absence of factor VIII is the cause of classic hemophilia. Factor VIII is encoded on the X chromosome, so the disease is much more prominent in males, because they have only one X chromosome. As wounds heal, clots must be removed. The principal agent for dissolving clots is an enzyme called plasmin, which is synthesized as the inactive zymogen called plasminogen. Plasminogen is activated by a number of proteases, the most important of which is tissue-type plasminogen activator (t-PA). tPA can be extremely effective in initiating the cascade to dissolve the unwanted blood clot involved in stroke or heart attack.