Leukaemia Section t(11;14)(q23;q24) Atlas of Genetics and Cytogenetics in Oncology and Haematology
... 14q24 DNA/RNA 29 exons, spans approximately 800 kb, ORF 2.3 kb. Protein 736 to 770 amino acids; 93-105 kDa; submembraneous scaffold protein that anchors glycine receptor to postsynaptic cytoskeletal elements through a putative microtubule binding motif. GPHN is also involved in molybdenum cofactor b ...
... 14q24 DNA/RNA 29 exons, spans approximately 800 kb, ORF 2.3 kb. Protein 736 to 770 amino acids; 93-105 kDa; submembraneous scaffold protein that anchors glycine receptor to postsynaptic cytoskeletal elements through a putative microtubule binding motif. GPHN is also involved in molybdenum cofactor b ...
britt-hannah - St Mary`s College Society
... challenging nature of some of the molecules studied, such as amyloid proteins. ...
... challenging nature of some of the molecules studied, such as amyloid proteins. ...
Peptide Bonds - Newcastle University
... Two amino acids joined together are called a dipeptide. The condensation reaction you have just seen can repeat so any more amino acids can add together in the same way, forming a long chain called a polypeptide. For each protein, the order of amino acid residues is specific and different – it is th ...
... Two amino acids joined together are called a dipeptide. The condensation reaction you have just seen can repeat so any more amino acids can add together in the same way, forming a long chain called a polypeptide. For each protein, the order of amino acid residues is specific and different – it is th ...
Protein structure homework: FAQ
... A: I did not tell you not to use the information button -- I warned you that it might be misreading and asked you to verify this information by comparing with what you see on the screen and what is in the actual pdb-file. That's why I suggested that you open this file with notepad or any other text ...
... A: I did not tell you not to use the information button -- I warned you that it might be misreading and asked you to verify this information by comparing with what you see on the screen and what is in the actual pdb-file. That's why I suggested that you open this file with notepad or any other text ...
No Slide Title
... • Group of residues with high contact density, number of contacts within domains is higher than the number of contacts between domains. • A stable unit of protein structure that can fold autonomously • A rigid body linked to other domains by flexible linkers • A portion of the protein that can be ac ...
... • Group of residues with high contact density, number of contacts within domains is higher than the number of contacts between domains. • A stable unit of protein structure that can fold autonomously • A rigid body linked to other domains by flexible linkers • A portion of the protein that can be ac ...
Pfam-A
... • Group of residues with high contact density, number of contacts within domains is higher than the number of contacts between domains. • A stable unit of protein structure that can fold autonomously • A rigid body linked to other domains by flexible linkers • A portion of the protein that can be ac ...
... • Group of residues with high contact density, number of contacts within domains is higher than the number of contacts between domains. • A stable unit of protein structure that can fold autonomously • A rigid body linked to other domains by flexible linkers • A portion of the protein that can be ac ...
Statistical Analysis Using Scaffold - Proteome Software
... • Name and size of the database searched (Swisprot or NCBI and the number of sequence entries) • Name and version of any additional software used for statistical analysis and an explanation of the analysis (Scaffold, #peptide requirements, probability settings) ...
... • Name and size of the database searched (Swisprot or NCBI and the number of sequence entries) • Name and version of any additional software used for statistical analysis and an explanation of the analysis (Scaffold, #peptide requirements, probability settings) ...
presentation source
... • Km is a measure of the affinity of an enzyme for its substrate – lower the Km the greater the affinity – Km= substrate concentration at 1/2 Vmax ...
... • Km is a measure of the affinity of an enzyme for its substrate – lower the Km the greater the affinity – Km= substrate concentration at 1/2 Vmax ...
Biological Data Mining
... called Post-synaptic receptors synaptic membrane vesicles and release their content into recognize them as a synaptic cleft signal and get activated which then transmit the signal on to other signaling components ...
... called Post-synaptic receptors synaptic membrane vesicles and release their content into recognize them as a synaptic cleft signal and get activated which then transmit the signal on to other signaling components ...
Abstract
... eye that is debilitating and highly recalcitrant to current therapies. A number of protein drugs are known to suppress inflammation that causes dry eye, but they have little or no effects when applied as eye drops because they are washed out quickly by the tear flow and therefore have little or no e ...
... eye that is debilitating and highly recalcitrant to current therapies. A number of protein drugs are known to suppress inflammation that causes dry eye, but they have little or no effects when applied as eye drops because they are washed out quickly by the tear flow and therefore have little or no e ...
Protein Expression and Purification
... • Search literature for precedent with similar/related protein • PepcDB Protein Expression Purification and Crystallization Database http://pepcdb.pdb.org/index.html • Protocols for cloning, expression, purification are stored and are searchable ...
... • Search literature for precedent with similar/related protein • PepcDB Protein Expression Purification and Crystallization Database http://pepcdb.pdb.org/index.html • Protocols for cloning, expression, purification are stored and are searchable ...
Sports Nutrition Advertisement Assignment
... 10lbs (210 Servings) 90% PROTEIN-Pure WPI is nature's purest and most effective protein Undenatured Whey Protein , High in BCAAs for lean muscle growth. Supports the body's immune system. High levels of essential amino acids. No Ace K or aspartame. Natural appetite suppressant. Pure WPI is nature's ...
... 10lbs (210 Servings) 90% PROTEIN-Pure WPI is nature's purest and most effective protein Undenatured Whey Protein , High in BCAAs for lean muscle growth. Supports the body's immune system. High levels of essential amino acids. No Ace K or aspartame. Natural appetite suppressant. Pure WPI is nature's ...
Anti-UBR1 Antibody
... protein and participates in the formation of a substrate-linked multi ubiquitin chain. It recognizes and binds to proteins bearing specific N-terminal residues that are destabilizing according to the N-end rule, leading to their ubiquitination and subsequent degradation (1). UBR1 is involved in panc ...
... protein and participates in the formation of a substrate-linked multi ubiquitin chain. It recognizes and binds to proteins bearing specific N-terminal residues that are destabilizing according to the N-end rule, leading to their ubiquitination and subsequent degradation (1). UBR1 is involved in panc ...
PPTX
... • This might be achieved by assigning confidence scores to different levels of the complex by which it collapses/expands… ...
... • This might be achieved by assigning confidence scores to different levels of the complex by which it collapses/expands… ...
Quantitative profiling of differentiation
... from human cells with less prefractionation would have resulted in the omission of a large number of peptides from CID because of the inability of the mass spectrometer to select for fragmentation of all or even most of the peptide ions coeluting in any chromatographic time window. The analysis by µ ...
... from human cells with less prefractionation would have resulted in the omission of a large number of peptides from CID because of the inability of the mass spectrometer to select for fragmentation of all or even most of the peptide ions coeluting in any chromatographic time window. The analysis by µ ...
Gene Section CPEB4 (cytoplasmic polyadenylation element binding protein 4)
... (PanIN - Pancreatic Intraepithelial Neoplasia), medium in high-grade PanINs and high in welldifferentiated PDAC. These data suggest that CPEB4 expression can be a prognostic factor in pancreatic carcinogenesis. Oncogenesis CPEB4 has been reported as a master gene involved in the reprogramming of can ...
... (PanIN - Pancreatic Intraepithelial Neoplasia), medium in high-grade PanINs and high in welldifferentiated PDAC. These data suggest that CPEB4 expression can be a prognostic factor in pancreatic carcinogenesis. Oncogenesis CPEB4 has been reported as a master gene involved in the reprogramming of can ...
Classwork - Biomonsters
... nucleotides (see board), draw a mini DNA molecule. Your drawing should meet the following requirements: • Contain 6 nucleotides total • Label A, T, C, and Gs • Label the 5’ and 3’ ends of each DNA strand ...
... nucleotides (see board), draw a mini DNA molecule. Your drawing should meet the following requirements: • Contain 6 nucleotides total • Label A, T, C, and Gs • Label the 5’ and 3’ ends of each DNA strand ...
Hello everyone
... immediate expulsion). The most prominent benefit of this method is that surface area is maximized to permit rapid protein breakdown / absorption by bacteria / mucosal epithelium. The primary drawback to this method is that it requires retaining your composure a bit more and lessening physical activi ...
... immediate expulsion). The most prominent benefit of this method is that surface area is maximized to permit rapid protein breakdown / absorption by bacteria / mucosal epithelium. The primary drawback to this method is that it requires retaining your composure a bit more and lessening physical activi ...
200 -- protein detection
... LABORATORY 2 -- DETECTION OF PROTEINS Background: Proteins may be detected by staining with the Biuret reagent. The Cu 2+ in the Biuret reagent reacts with peptide bonds in proteins to form a violet color. Since free amino acids do not have a peptide bond, they will not react with the Biuret reagent ...
... LABORATORY 2 -- DETECTION OF PROTEINS Background: Proteins may be detected by staining with the Biuret reagent. The Cu 2+ in the Biuret reagent reacts with peptide bonds in proteins to form a violet color. Since free amino acids do not have a peptide bond, they will not react with the Biuret reagent ...
Correlating heritable traits with specific gene products.
... emergence and biological functions of cytokines. The challenge in this genomic age is to detect novel molecules--otherwise buried in the dark recesses of sequence databases--that may have weak or unapparent ties to existing cytokine groups; we can do this best with computational techniques that are ...
... emergence and biological functions of cytokines. The challenge in this genomic age is to detect novel molecules--otherwise buried in the dark recesses of sequence databases--that may have weak or unapparent ties to existing cytokine groups; we can do this best with computational techniques that are ...
Choose My Plate
... Go to the website choosemyplate.gov. From the main webpage click on the green tab in the upper left corner labeled MyPlate. There should be a list of the 6 major food groups along the left portion of the page; find the answers to the following questions by navigating through the information found wi ...
... Go to the website choosemyplate.gov. From the main webpage click on the green tab in the upper left corner labeled MyPlate. There should be a list of the 6 major food groups along the left portion of the page; find the answers to the following questions by navigating through the information found wi ...
Toober variations
... photo. 2. Unfold the protein and then ask them to re-fold the toober into the original shape. 3. Check the refolded protein against the photo of the native structure. ...
... photo. 2. Unfold the protein and then ask them to re-fold the toober into the original shape. 3. Check the refolded protein against the photo of the native structure. ...
Poster
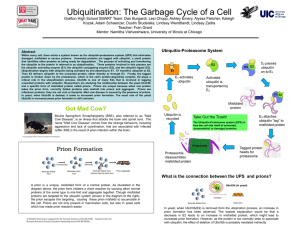
... Within every cell, there exists a system known as the ubiquitin-proteasome system (UPS) that eliminates damaged, misfolded or excess proteins. Unwanted proteins are tagged with ubiquitin, a small protein that identifies other proteins as being ready for degradation. The process of activating and tra ...
... Within every cell, there exists a system known as the ubiquitin-proteasome system (UPS) that eliminates damaged, misfolded or excess proteins. Unwanted proteins are tagged with ubiquitin, a small protein that identifies other proteins as being ready for degradation. The process of activating and tra ...
DNA, RNA, Proteins
... 3. Proteins: Biopolymers interconnected with peptide bonds Function: most important molecules of the cell. Highly diverse functions - structure, chemical catalysis energy transduction, ...
... 3. Proteins: Biopolymers interconnected with peptide bonds Function: most important molecules of the cell. Highly diverse functions - structure, chemical catalysis energy transduction, ...
Proteomics

Proteomics is the large-scale study of proteins, particularly their structures and functions. Proteins are vital parts of living organisms, as they are the main components of the physiological metabolic pathways of cells. The term proteomics was first coined in 1997 to make an analogy with genomics, the study of the genome. The word proteome is a portmanteau of protein and genome, and was coined by Marc Wilkins in 1994 while working on the concept as a PhD student.The proteome is the entire set of proteins, produced or modified by an organism or system. This varies with time and distinct requirements, or stresses, that a cell or organism undergoes. Proteomics is an interdisciplinary domain formed on the basis of the research and development of the Human Genome Project; it is also emerging scientific research and exploration of proteomes from the overall level of intracellular protein composition, structure, and its own unique activity patterns. It is an important component of functional genomics.While proteomics generally refers to the large-scale experimental analysis of proteins, it is often specifically used for protein purification and mass spectrometry.