Supplementary Information (doc 132K)
... After separating the final samples into three subpopulation-specific strata (EU, AJ, SA), an additional set of QC analyses were undertaken within each subpopulation to optimize casecontrol matching and to remove remaining poorly performing samples and SNPs (Supplementary Figure S1). First, samples ...
... After separating the final samples into three subpopulation-specific strata (EU, AJ, SA), an additional set of QC analyses were undertaken within each subpopulation to optimize casecontrol matching and to remove remaining poorly performing samples and SNPs (Supplementary Figure S1). First, samples ...
Functional SNPs in the SCGB3A2 promoter are
... for linkage at D5s436 on chromosome 5q31. When four additional markers around D5s436 were used, a maximum two-point LOD score of 4.31 and a maximum multipoint LOD score of 4.12 were obtained for marker D5s2090 (11). Interestingly, from a dataset of 123 Japanese sibling pairs, the 5q31 locus was also ...
... for linkage at D5s436 on chromosome 5q31. When four additional markers around D5s436 were used, a maximum two-point LOD score of 4.31 and a maximum multipoint LOD score of 4.12 were obtained for marker D5s2090 (11). Interestingly, from a dataset of 123 Japanese sibling pairs, the 5q31 locus was also ...
Dissecting the genetics variation of aggressive behaviour in
... eliminated due to quality control in the call rate or because they were monomorphic. A further 13364 (21.4 %) SNP were eliminated because their minor allele frequency was less than 0.05 or because their 12 value exceeded 3.89 when testing their genotype frequencies for deviation of what it is expe ...
... eliminated due to quality control in the call rate or because they were monomorphic. A further 13364 (21.4 %) SNP were eliminated because their minor allele frequency was less than 0.05 or because their 12 value exceeded 3.89 when testing their genotype frequencies for deviation of what it is expe ...
Here is the Original File
... method of looking through a vast amount of genomic data to determine whether any part or parts of the genome contain single nucleotide polymorphism (SNP) variants associated with a specific trait. Due to epistasis, in which the effects of one gene are modified by one or several other genes1, there h ...
... method of looking through a vast amount of genomic data to determine whether any part or parts of the genome contain single nucleotide polymorphism (SNP) variants associated with a specific trait. Due to epistasis, in which the effects of one gene are modified by one or several other genes1, there h ...
What is bioinformatics? - The British Association of Sport and
... The human and many other genomes have now been sequenced and this data has been deposited online. In addition, there is a wealth of information on genes and their products on networked computers. Numerous programmes that allow you to analyse this data do also exist. Most of this data is freely acces ...
... The human and many other genomes have now been sequenced and this data has been deposited online. In addition, there is a wealth of information on genes and their products on networked computers. Numerous programmes that allow you to analyse this data do also exist. Most of this data is freely acces ...
Ross - Tree Improvement Program
... • Select regions to screen for possible SNPs • Amplify and sequence target regions from test population samples • Analyze data for SNP markers that will be useful for fingerprinting ...
... • Select regions to screen for possible SNPs • Amplify and sequence target regions from test population samples • Analyze data for SNP markers that will be useful for fingerprinting ...
Fine scale mapping
... f(A,U|x,M) = f(A|x,M) f(U|h) The likelihood, f(U|h), depends only on population SNP haplotype frequencies, h. For many SNPs, the number of possible haplotypes is large, so frequencies are parameterised in terms of allele frequencies and first-order LD between pairs of adjacent loci. ...
... f(A,U|x,M) = f(A|x,M) f(U|h) The likelihood, f(U|h), depends only on population SNP haplotype frequencies, h. For many SNPs, the number of possible haplotypes is large, so frequencies are parameterised in terms of allele frequencies and first-order LD between pairs of adjacent loci. ...
Slide 1
... expressivity), let alone genetic and environmental modifiers contributing to it (e.g., the influence of pregnancy and smoking on carriers of BRCA1/2). - Also, missing from a majority: “gene-environment interactions and the potential influence of these interactions on disease” (from L. Doyle, ...
... expressivity), let alone genetic and environmental modifiers contributing to it (e.g., the influence of pregnancy and smoking on carriers of BRCA1/2). - Also, missing from a majority: “gene-environment interactions and the potential influence of these interactions on disease” (from L. Doyle, ...
Review of Intestinal Drug Absorption and Intestinal
... • Pharmacology + Genomics = Pharmacogenomics • The study of how an individual’s genetic inheritance affects the body’s response to drugs ...
... • Pharmacology + Genomics = Pharmacogenomics • The study of how an individual’s genetic inheritance affects the body’s response to drugs ...
Genome-Wide Prediction of Functional Gene
... in complex traits is to assume functional interactions between genes. Therefore we searched two mammalian genomes to identify potential epistatic interactions by looking for co-adapted genes marked by excess two-locus genetic differentiation between populations/lineages using publicly available SNP ...
... in complex traits is to assume functional interactions between genes. Therefore we searched two mammalian genomes to identify potential epistatic interactions by looking for co-adapted genes marked by excess two-locus genetic differentiation between populations/lineages using publicly available SNP ...
of genome-wide association studies
... Andrew P. Morris, PhD, Professor of Statistical Genetics, Wellcome Trust Senior Research Fellow in Basic Biomedical Science, University of Liverpool, Liverpool, UK Reedik Mägi, PhD, Senior Research Fellow, Head of Bioinformatics workgroup, Estonian Genome Center, University of Tartu, Tartu, Estonia ...
... Andrew P. Morris, PhD, Professor of Statistical Genetics, Wellcome Trust Senior Research Fellow in Basic Biomedical Science, University of Liverpool, Liverpool, UK Reedik Mägi, PhD, Senior Research Fellow, Head of Bioinformatics workgroup, Estonian Genome Center, University of Tartu, Tartu, Estonia ...
Ohio State Talk, October 2004
... then the data we would see would simply be the unordered set (A,a,B,b) • However, this is also consistent with a different diplotype, namely Hdip = {(am,Bm), (Af,bf)} • Note that the number of copies of the (a,b) haplotype differs in these two cases • The true diploid = haplotype pair is missing ...
... then the data we would see would simply be the unordered set (A,a,B,b) • However, this is also consistent with a different diplotype, namely Hdip = {(am,Bm), (Af,bf)} • Note that the number of copies of the (a,b) haplotype differs in these two cases • The true diploid = haplotype pair is missing ...
Genetic Update Conferences - 2002 - yhs
... 'Variants' - differences among individuals 'Normal' or 'Wild Type' - most common variants 'Mutation' - DNA variant that is pathologic / causes disease "We are all Mutants" E. Person to Person Genomic Sequence - 99.6% identical DNA Sequence 0.4% different = 24,000,000 bp in the genome - 24,000,000var ...
... 'Variants' - differences among individuals 'Normal' or 'Wild Type' - most common variants 'Mutation' - DNA variant that is pathologic / causes disease "We are all Mutants" E. Person to Person Genomic Sequence - 99.6% identical DNA Sequence 0.4% different = 24,000,000 bp in the genome - 24,000,000var ...
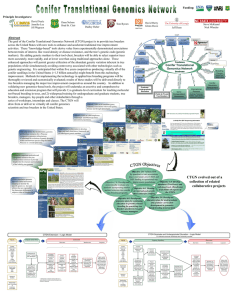
Objective 2.0
... across the United States with new tools to enhance and accelerate traditional tree improvement activities. These “knowledge-based” tools derive value from experimentally demonstrated associations between traits of interest, like wood density or disease resistance, and the tree’s genetic code (geneti ...
... across the United States with new tools to enhance and accelerate traditional tree improvement activities. These “knowledge-based” tools derive value from experimentally demonstrated associations between traits of interest, like wood density or disease resistance, and the tree’s genetic code (geneti ...
Exploring the Importance of Single Nucleotide Polymorphisms of
... Based on the results, all three of the odds ratios, with confidence intervals, included one. If an odds ratio is one the data set is not statistically significant because including one means that there is not a haplotype that is more or less likely to be a haplotype in a case than in a control. This ...
... Based on the results, all three of the odds ratios, with confidence intervals, included one. If an odds ratio is one the data set is not statistically significant because including one means that there is not a haplotype that is more or less likely to be a haplotype in a case than in a control. This ...
A Genome-Wide Association Study of Inbred Rat Strains
... genes related to Parkinson's Disease1, Type II Diabetes2, human height variation3 and more. These studies share a number of common characteristics. The studies are done using human subjects. Each individual is measured for both genotype and phenotype. The phenotypes may be quantitative measurements ...
... genes related to Parkinson's Disease1, Type II Diabetes2, human height variation3 and more. These studies share a number of common characteristics. The studies are done using human subjects. Each individual is measured for both genotype and phenotype. The phenotypes may be quantitative measurements ...
Modifier genes in Huntington`s desease - Ruhr
... Altogether, this project investigated and identified genetic variations associated with mitochondrial function and biogenesis. The interplay between mitochondrial and nuclear encoded genes in modification of AO has revealed novel aspects concerning the genetic basis of mitochondrial failure in HD. T ...
... Altogether, this project investigated and identified genetic variations associated with mitochondrial function and biogenesis. The interplay between mitochondrial and nuclear encoded genes in modification of AO has revealed novel aspects concerning the genetic basis of mitochondrial failure in HD. T ...
NIHMS27833-supplement-1 - TARA
... That a number of SNPs in Supplementary Table 5 do not show association in our sample does not invalidate the original findings. There are some caveats to our analysis; for example, not all SNPs were directly genotyped in our GWAS. An attempt was made to identify proxy SNPs, but for some the LD betw ...
... That a number of SNPs in Supplementary Table 5 do not show association in our sample does not invalidate the original findings. There are some caveats to our analysis; for example, not all SNPs were directly genotyped in our GWAS. An attempt was made to identify proxy SNPs, but for some the LD betw ...
Nyssa Fox
... schizophrenia, researchers first had to determine whether the dysfunction was due to a genetic or environmental factor. Mental health professionals had noticed that schizophrenia tends to run in families, but research found that the heritability factor had a large range, between 41-87%.5 To determin ...
... schizophrenia, researchers first had to determine whether the dysfunction was due to a genetic or environmental factor. Mental health professionals had noticed that schizophrenia tends to run in families, but research found that the heritability factor had a large range, between 41-87%.5 To determin ...
NUS Presentation Title 2006
... Shared and unique influences upon mRNA • What proportion of an individual gene’s variation can be explained by shared influences? • Cis acting variations 15-40% ...
... Shared and unique influences upon mRNA • What proportion of an individual gene’s variation can be explained by shared influences? • Cis acting variations 15-40% ...
Chromosome 21 Scan in Down Syndrome Reveals DSCAM as a
... Genotyping was performed using R 2.15.2 software by taking the following steps: (i) intensity extraction (3 intensities per SNP allele using packages pd.genomewide.6 and oligoClasses [9,10]), (ii) normalization (i.e ratio of the mean intensity obtained for one of the SNP allele divided by the sum of ...
... Genotyping was performed using R 2.15.2 software by taking the following steps: (i) intensity extraction (3 intensities per SNP allele using packages pd.genomewide.6 and oligoClasses [9,10]), (ii) normalization (i.e ratio of the mean intensity obtained for one of the SNP allele divided by the sum of ...
Examination of G72 and D-amino-acid oxidase
... with BPAD (P¼0.032), but the global P-value did not reach significance (P¼0.08) (Table 2). It should be noted that M12 and M15 as well as M23 and M24 show a high degree of intermarker LD (Table 3) in our study. The three SNPs in the DAAO gene were associated with schizophrenia, but failed to reach s ...
... with BPAD (P¼0.032), but the global P-value did not reach significance (P¼0.08) (Table 2). It should be noted that M12 and M15 as well as M23 and M24 show a high degree of intermarker LD (Table 3) in our study. The three SNPs in the DAAO gene were associated with schizophrenia, but failed to reach s ...
GENES AND CHROMOSOMES
... 2. gametes carry one allele or the other, but not both B. when two pairs of alternate alleles carried on two pairs of homologs 1. homologs separate during meiosis I 2. chromatids separate during meiosis II 3. alleles assort independently II. Linkage A. definition of linked genes: B. relation to Mend ...
... 2. gametes carry one allele or the other, but not both B. when two pairs of alternate alleles carried on two pairs of homologs 1. homologs separate during meiosis I 2. chromatids separate during meiosis II 3. alleles assort independently II. Linkage A. definition of linked genes: B. relation to Mend ...
GLYPHOSATE RESISTANCE Background / Problem
... Synonymous substitution rate can be used to set neutral expectation for nonsynonymous rate dS is the relative rate of synonymous mutations ...
... Synonymous substitution rate can be used to set neutral expectation for nonsynonymous rate dS is the relative rate of synonymous mutations ...
Tag SNP

A tag SNP is a representative single nucleotide polymorphism (SNP) in a region of the genome with high linkage disequilibrium that represents a group of SNPs called a haplotype. It is possible to identify genetic variation and association to phenotypes without genotyping every SNP in a chromosomal region. This reduces the expense and time of mapping genome areas associated with disease, since it eliminates the need to study every individual SNP. Tag SNPs are useful in whole-genome SNP association studies in which hundreds of thousands of SNPs across the entire genome are genotyped.