proteins
... – a central carbon atom (called the "alpha carbon"), with four different chemical groups attached to it: -a hydrogen atom -a basic amino group -an acidic carboxyl group -a variable "R" group (or side chain) ...
... – a central carbon atom (called the "alpha carbon"), with four different chemical groups attached to it: -a hydrogen atom -a basic amino group -an acidic carboxyl group -a variable "R" group (or side chain) ...
aLFQ: an R-package for estimating absolute protein quantities from
... The current gold standard for LC-MS/MS–based absolute protein quantification is the use of stable isotope-labeled standard (SIS) peptides or proteins in precisely determined concentrations (Brun et al., 2009). These standards are spiked into the biological sample of interest and the absolute concent ...
... The current gold standard for LC-MS/MS–based absolute protein quantification is the use of stable isotope-labeled standard (SIS) peptides or proteins in precisely determined concentrations (Brun et al., 2009). These standards are spiked into the biological sample of interest and the absolute concent ...
Alzheimer`s - Science Nutshell
... be obtained from alternatively spliced mRNA. These mutations lead to an amyloidogenic form of beta amyloid peptide (A42/43), which is longer in length. The most severe mutations cause large a decrease in A40, and a large increase in A42. A40 doesn’t bind to more than 3 other A40 proteins, where ...
... be obtained from alternatively spliced mRNA. These mutations lead to an amyloidogenic form of beta amyloid peptide (A42/43), which is longer in length. The most severe mutations cause large a decrease in A40, and a large increase in A42. A40 doesn’t bind to more than 3 other A40 proteins, where ...
Bioc 462a Lecture Notes
... Total protein was measured by determining the absorbance at 280 nm. In order to follow the fatty acid-binding proteins, they were labeled by binding radioactive fatty acids (CPM=counts per minute - gray shading). The purity of each peak was assessed using SDS-PAGE (insert). There are two, nearly pur ...
... Total protein was measured by determining the absorbance at 280 nm. In order to follow the fatty acid-binding proteins, they were labeled by binding radioactive fatty acids (CPM=counts per minute - gray shading). The purity of each peak was assessed using SDS-PAGE (insert). There are two, nearly pur ...
No Slide Title
... strongly, which one weakly, and which ones not at all. Since we also have NOE and 3J coupling data, we can try to see if these hydrogen bonded amides match with regions that we identified previously as a-helices, b-sheets, or b-turns. • If we can do this, then, and ONLY then, we can use a H-bond con ...
... strongly, which one weakly, and which ones not at all. Since we also have NOE and 3J coupling data, we can try to see if these hydrogen bonded amides match with regions that we identified previously as a-helices, b-sheets, or b-turns. • If we can do this, then, and ONLY then, we can use a H-bond con ...
Ser-Ala-Trp
... D. Exhibit a tightly coiled polypeptide main chain from which the R groups extend outward. ...
... D. Exhibit a tightly coiled polypeptide main chain from which the R groups extend outward. ...
Purified Sp1 protein
... Buffer Contents: 2 µg of Purified Sp1 protein and Dilution Buffer AM1 (20 mM TrisCl (pH 8), 20% glycerol, 100 mM KCl, 1 mM DTT and 0.2 mM EDTA). Protein is supplied at 0.08 µg/µl in Dilution Buffer AM1. Background: Sp1 (specificity protein 1) is a human transcription factor involved in gene expressi ...
... Buffer Contents: 2 µg of Purified Sp1 protein and Dilution Buffer AM1 (20 mM TrisCl (pH 8), 20% glycerol, 100 mM KCl, 1 mM DTT and 0.2 mM EDTA). Protein is supplied at 0.08 µg/µl in Dilution Buffer AM1. Background: Sp1 (specificity protein 1) is a human transcription factor involved in gene expressi ...
Peptidomimetics- A definition
... considered (hormones, peptides and transmitters). Peptidomimetic molecules are regular organic molecules which are conceived to mimic the structure of an endogenous peptide. These peptidomimetics are valuable because peptides can rarely be developed as drugs. In general peptides are: •Biologically u ...
... considered (hormones, peptides and transmitters). Peptidomimetic molecules are regular organic molecules which are conceived to mimic the structure of an endogenous peptide. These peptidomimetics are valuable because peptides can rarely be developed as drugs. In general peptides are: •Biologically u ...
Lecture 7: Protein Sorting
... Nuclear import receptors bind to nuclear porins and nuclear localization signal of cargo protein ...
... Nuclear import receptors bind to nuclear porins and nuclear localization signal of cargo protein ...
Dynabeads® for protein complex isolation
... 9. Hayes, M.J. et al. (2006) Early mitotic degradation of Nek2A depends on Cdc20-independent interaction with the APC/C. Nature Cell Biology 8(6) : 607–614. 10. Kantardzhieva, A. et al. (2006) MPP3 is recruited to the MPP5 protein scaffold at the retinal outer limiting membrane. FEBS Journal 273: 11 ...
... 9. Hayes, M.J. et al. (2006) Early mitotic degradation of Nek2A depends on Cdc20-independent interaction with the APC/C. Nature Cell Biology 8(6) : 607–614. 10. Kantardzhieva, A. et al. (2006) MPP3 is recruited to the MPP5 protein scaffold at the retinal outer limiting membrane. FEBS Journal 273: 11 ...
Getting things where they need to go: Protein Targeting
... Light driven pumps (primarily bacterial) ...
... Light driven pumps (primarily bacterial) ...
Fundamentals of protein structure
... •Generally proteins are soluble in water, except the membrane proteins which are hydrophobic •Absorb light in the UV range. Maximum absorption at 280nm due to aromatic a.a •Have a specific Isoelectric pH [pI]. Positively charged below PI, and negatively charged above pI •Proteins are charged molecul ...
... •Generally proteins are soluble in water, except the membrane proteins which are hydrophobic •Absorb light in the UV range. Maximum absorption at 280nm due to aromatic a.a •Have a specific Isoelectric pH [pI]. Positively charged below PI, and negatively charged above pI •Proteins are charged molecul ...
In Depth Analysis of Protein Amino Acid Sequence and PTMs with
... • Peptides from the set of confident proteins are “modified” in-silico by trying all possible modifications in UNIMOD. • Speed up by de novo tags PeaksPTM: Mass spectrometry-based identification of peptides with unspecified modifications. Journal of Proteome Research 10.7 (2011) : 2930-2936 ...
... • Peptides from the set of confident proteins are “modified” in-silico by trying all possible modifications in UNIMOD. • Speed up by de novo tags PeaksPTM: Mass spectrometry-based identification of peptides with unspecified modifications. Journal of Proteome Research 10.7 (2011) : 2930-2936 ...
Summer 1997, First Half, Dr
... Discuss the structure of the -helix in proteins and give several examples where this structural motif is found. ...
... Discuss the structure of the -helix in proteins and give several examples where this structural motif is found. ...
Facts and Fallacies
... Order of de Novo and DB • Better conduct de novo on all spectra. – De novo not slow, and computing is cheap. – De novo provides independent validation for DB result. # consensus AA (de novo vs. DB search) ...
... Order of de Novo and DB • Better conduct de novo on all spectra. – De novo not slow, and computing is cheap. – De novo provides independent validation for DB result. # consensus AA (de novo vs. DB search) ...
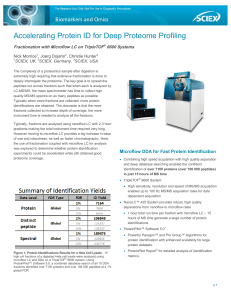
Accelerating Protein ID for Deep Proteome Profiling
... The complexity of a proteomics sample after digestion is extremely high requiring that extensive fractionation is done to deeply interrogate the proteome. The key goal is to spread the peptides out across fractions such that when each is analyzed by LC-MS/MS, the mass spectrometer has time to collec ...
... The complexity of a proteomics sample after digestion is extremely high requiring that extensive fractionation is done to deeply interrogate the proteome. The key goal is to spread the peptides out across fractions such that when each is analyzed by LC-MS/MS, the mass spectrometer has time to collec ...
Health significance of protein
... diet has to supply any essential amino acids, otherwise the body breaks down its own protein (e.g. muscle protein) to obtain them. In other words, the more essential amino acids the protein provides, the higher its quality. ...
... diet has to supply any essential amino acids, otherwise the body breaks down its own protein (e.g. muscle protein) to obtain them. In other words, the more essential amino acids the protein provides, the higher its quality. ...
Additional file 6
... All the identified proteins of the non-redundant, high-confidence dataset of glomerulus proteome consisting of 1,817 unique proteins representing 1,478 unique genes were analyzed based onGene Ontology (GO) Cellular Component (Panel A) and GO Molecular Function (Panel B) vocabularies using PANTHER ve ...
... All the identified proteins of the non-redundant, high-confidence dataset of glomerulus proteome consisting of 1,817 unique proteins representing 1,478 unique genes were analyzed based onGene Ontology (GO) Cellular Component (Panel A) and GO Molecular Function (Panel B) vocabularies using PANTHER ve ...
lecture 4
... cavities and assist protein folding. The eukaryotic cytosolic chaperonin is involved in folding actins and tubulins. Prefoldin. Hexameric molecular chaperone also involved in actin and tubulin biogenesis. Its existence was not known when the Cell paper was published in 1997 (it was discovered in 199 ...
... cavities and assist protein folding. The eukaryotic cytosolic chaperonin is involved in folding actins and tubulins. Prefoldin. Hexameric molecular chaperone also involved in actin and tubulin biogenesis. Its existence was not known when the Cell paper was published in 1997 (it was discovered in 199 ...
Ms. Robyn Klemptner
... Compare enrichment of in-gel digested proteins to proteins in solution – efficiency of dendrimer-based enrichments. Compare genomic, proteomic and metabolomic data. ...
... Compare enrichment of in-gel digested proteins to proteins in solution – efficiency of dendrimer-based enrichments. Compare genomic, proteomic and metabolomic data. ...
The cost of life is energy.
... • How is this like the lock and key model of enzyme function? • How would toothpickase function if you had to break toothpicks with your hands in the Arctic Ocean? Under a broiler? Submerged in a vat of acid? • What effect would it have on toothpickase action if there were a high concentration of to ...
... • How is this like the lock and key model of enzyme function? • How would toothpickase function if you had to break toothpicks with your hands in the Arctic Ocean? Under a broiler? Submerged in a vat of acid? • What effect would it have on toothpickase action if there were a high concentration of to ...
Poster presentations
... (A printable MSWord version of this page) Aharonovsky Elik (University of Haifa) Protein sequence modules and their relation to closed loops. Akabayov Barak, Irit Sagi (Weizmann Institute) The metal ion in the RNA helicase DbpA acts as a single atom turnover switch during catalysis. Ben-Shimon Avi ( ...
... (A printable MSWord version of this page) Aharonovsky Elik (University of Haifa) Protein sequence modules and their relation to closed loops. Akabayov Barak, Irit Sagi (Weizmann Institute) The metal ion in the RNA helicase DbpA acts as a single atom turnover switch during catalysis. Ben-Shimon Avi ( ...
Top down - The Fenyo Lab
... BLACK: duplicated experiments, FAl treated cells (then ground) SCORE: Log Ion Current / Log protein abundance ...
... BLACK: duplicated experiments, FAl treated cells (then ground) SCORE: Log Ion Current / Log protein abundance ...
Sonac introduces new natural products for aqua feed
... conventional sources of cholesterol, Sonac recently developed Phosterol. This hydrolyzed protein of animal origin is produced in accordance with all relevant EU regulations and is readily available from renewable natural sources. The uniqueness of Phosterol lays in the natural combination of cholest ...
... conventional sources of cholesterol, Sonac recently developed Phosterol. This hydrolyzed protein of animal origin is produced in accordance with all relevant EU regulations and is readily available from renewable natural sources. The uniqueness of Phosterol lays in the natural combination of cholest ...
Histone Demethylation by A Family of JmjC Domain
... Figure S2. Schematic representation of the steps used in purifying the demethylase activity from HeLa cells. Numbers represent the salt concentrations (mM) at which the histone demethylase activity elutes from the column. Figure S3. Comparison of the JHDM1 family of proteins. a. Diagrammatic represe ...
... Figure S2. Schematic representation of the steps used in purifying the demethylase activity from HeLa cells. Numbers represent the salt concentrations (mM) at which the histone demethylase activity elutes from the column. Figure S3. Comparison of the JHDM1 family of proteins. a. Diagrammatic represe ...
Protein mass spectrometry

Protein mass spectrometry refers to the application of mass spectrometry to the study of proteins. Mass spectrometry is an important emerging method for the characterization of proteins. The two primary methods for ionization of whole proteins are electrospray ionization (ESI) and matrix-assisted laser desorption/ionization (MALDI). In keeping with the performance and mass range of available mass spectrometers, two approaches are used for characterizing proteins. In the first, intact proteins are ionized by either of the two techniques described above, and then introduced to a mass analyzer. This approach is referred to as ""top-down"" strategy of protein analysis. In the second, proteins are enzymatically digested into smaller peptides using a protease such as trypsin. Subsequently these peptides are introduced into the mass spectrometer and identified by peptide mass fingerprinting or tandem mass spectrometry. Hence, this latter approach (also called ""bottom-up"" proteomics) uses identification at the peptide level to infer the existence of proteins.Whole protein mass analysis is primarily conducted using either time-of-flight (TOF) MS, or Fourier transform ion cyclotron resonance (FT-ICR). These two types of instrument are preferable here because of their wide mass range, and in the case of FT-ICR, its high mass accuracy. Mass analysis of proteolytic peptides is a much more popular method of protein characterization, as cheaper instrument designs can be used for characterization. Additionally, sample preparation is easier once whole proteins have been digested into smaller peptide fragments. The most widely used instrument for peptide mass analysis are the MALDI time-of-flight instruments as they permit the acquisition of peptide mass fingerprints (PMFs) at high pace (1 PMF can be analyzed in approx. 10 sec). Multiple stage quadrupole-time-of-flight and the quadrupole ion trap also find use in this application.