Introduction to Biomolecular Structure
... • Above pH 7: lower [H+] basic • Cellular pH is approximately 7.2-7.4. ...
... • Above pH 7: lower [H+] basic • Cellular pH is approximately 7.2-7.4. ...
Lesson Overview
... Some transcription factors enhance transcription by opening up tightly packed chromatin. Others help attract RNA polymerase. Still others block access to certain genes. In most cases, multiple transcription factors must bind before RNA polymerase is able to attach to the promoter region and start tr ...
... Some transcription factors enhance transcription by opening up tightly packed chromatin. Others help attract RNA polymerase. Still others block access to certain genes. In most cases, multiple transcription factors must bind before RNA polymerase is able to attach to the promoter region and start tr ...
Slide 1
... Some transcription factors enhance transcription by opening up tightly packed chromatin. Others help attract RNA polymerase. Still others block access to certain genes. In most cases, multiple transcription factors must bind before RNA polymerase is able to attach to the promoter region and start tr ...
... Some transcription factors enhance transcription by opening up tightly packed chromatin. Others help attract RNA polymerase. Still others block access to certain genes. In most cases, multiple transcription factors must bind before RNA polymerase is able to attach to the promoter region and start tr ...
BACTERIAL GENETICS
... • RNA -ribose instead of deoxyribose and uracil instead of thymine • Central dogma of molecular biology • DNA transcription RNA ribosomes polypeptide • mRNA,tRNA,rRNA. • Genetic information is stored in DNA as code. ...
... • RNA -ribose instead of deoxyribose and uracil instead of thymine • Central dogma of molecular biology • DNA transcription RNA ribosomes polypeptide • mRNA,tRNA,rRNA. • Genetic information is stored in DNA as code. ...
大碩102研究所全真模擬考試試題
... (B) It will lead to a tighter association of histone with DNA, resulting in reduced transcription. (C) It will have no effect on the interaction of DNA with histones. (D) There will be a complete repression of transcription. (E) None of the choices are correct. 28. If Hershey and Chase found S35 in ...
... (B) It will lead to a tighter association of histone with DNA, resulting in reduced transcription. (C) It will have no effect on the interaction of DNA with histones. (D) There will be a complete repression of transcription. (E) None of the choices are correct. 28. If Hershey and Chase found S35 in ...
Southern transfer
... eukaryotes are still some way beyond the capability of the current technology. Gene location on these larger DNA molecules can ,however, be achieved by in situ hybridization, which has the added advantage : 1- Not only identifying which chromosome a gene lies on, 2-but also providing information on ...
... eukaryotes are still some way beyond the capability of the current technology. Gene location on these larger DNA molecules can ,however, be achieved by in situ hybridization, which has the added advantage : 1- Not only identifying which chromosome a gene lies on, 2-but also providing information on ...
Mathematical Challenges from Genomics and Molecular Biology
... agricultural, or environmental studies. In addition, it may be useful for comparative studies with related organisms. 2. Identify the genes and determine the functions of the proteins they encode. This process is essential, since without it a sequenced genome is merely a meaningless jumble of A’s, C ...
... agricultural, or environmental studies. In addition, it may be useful for comparative studies with related organisms. 2. Identify the genes and determine the functions of the proteins they encode. This process is essential, since without it a sequenced genome is merely a meaningless jumble of A’s, C ...
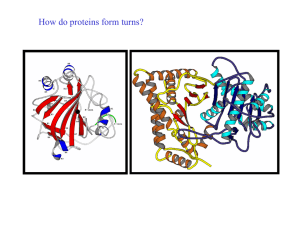
How do proteins form turns? - UF Macromolecular Structure Group
... A reverse turn is region of the polypeptide having a hydrogen bond from one main chain carbonyl oxygen to the main chain N-H group 3 residues along the chain (i.e. O(i) to N(i+3)) Helical regions are excluded from this definition (see later) Reverse turns are very abundant in globular proteins and g ...
... A reverse turn is region of the polypeptide having a hydrogen bond from one main chain carbonyl oxygen to the main chain N-H group 3 residues along the chain (i.e. O(i) to N(i+3)) Helical regions are excluded from this definition (see later) Reverse turns are very abundant in globular proteins and g ...
G W B enes at
... that are matched to a person’s unique genetic profile. Researchers believe that these customized drugs will be much less likely than current medicines to cause side effects. RNA interference (RNAi) is a technique that takes advantage of the ability of small RNAs to modify gene expression. In the fut ...
... that are matched to a person’s unique genetic profile. Researchers believe that these customized drugs will be much less likely than current medicines to cause side effects. RNA interference (RNAi) is a technique that takes advantage of the ability of small RNAs to modify gene expression. In the fut ...
PDF file of the lecture on "Gene Transfer"
... than the ability to insert copies of themselves into the bacterial chromosome. • IS form copies of themselves and the copies move into other areas of the chromosome. • They can interrupt the coding ...
... than the ability to insert copies of themselves into the bacterial chromosome. • IS form copies of themselves and the copies move into other areas of the chromosome. • They can interrupt the coding ...
video slide - CARNES AP BIO
... polypeptide, depending on which segments are treated as exons during RNA splicing • Such variations are called alternative RNA splicing • Because of alternative splicing, the number of different proteins an organism can produce is much greater than its number of genes ...
... polypeptide, depending on which segments are treated as exons during RNA splicing • Such variations are called alternative RNA splicing • Because of alternative splicing, the number of different proteins an organism can produce is much greater than its number of genes ...
12–3 RNA and Protein Synthesis
... Transcription RNA molecules are produced by copying part of a nucleotide sequence of DNA into a complementary sequence in RNA. This process is called transcription. Transcription requires another enzyme, RNA polymerase. ...
... Transcription RNA molecules are produced by copying part of a nucleotide sequence of DNA into a complementary sequence in RNA. This process is called transcription. Transcription requires another enzyme, RNA polymerase. ...
P site
... The segment of DNA that contains genetic information for a specific protein is uncoupled by RNA polymerase. Free ribonucleotides (ATP, UTP, CTP and GTP) are automatically attracted by the exposed bases on one polynucleotide strand (the sense strand) which contains the genetic information. New nucleo ...
... The segment of DNA that contains genetic information for a specific protein is uncoupled by RNA polymerase. Free ribonucleotides (ATP, UTP, CTP and GTP) are automatically attracted by the exposed bases on one polynucleotide strand (the sense strand) which contains the genetic information. New nucleo ...
Protein synthesis test review key
... The process of protein synthesis begins in the nucleus. Transcription occurs in the nucleus. DNA is transcribed into mRNA. Base pairs match up and create an mRNA strand. The mRNA then travels to the ribosome where translation occurs. tRNA brings amino acids to the ribosome based on the mRNA strand. ...
... The process of protein synthesis begins in the nucleus. Transcription occurs in the nucleus. DNA is transcribed into mRNA. Base pairs match up and create an mRNA strand. The mRNA then travels to the ribosome where translation occurs. tRNA brings amino acids to the ribosome based on the mRNA strand. ...
Honors_Genetics_B_Student_Notes
... Mutation- any change in a cell’s DNA sequence - Some mutations are harmful, some are beneficial and some have no effect on the organism ...
... Mutation- any change in a cell’s DNA sequence - Some mutations are harmful, some are beneficial and some have no effect on the organism ...
Watson, Crick and Wilkins
... the discovery of restriction enzymes and their application to problems of molecular genetics” Arber discovered these enzymes in the early 1960s when he analyzed an apparently obscure phenomenon in bacteria, discovered 10 years earlier by Bertani and Weigle, called host-controlled modification. Arber ...
... the discovery of restriction enzymes and their application to problems of molecular genetics” Arber discovered these enzymes in the early 1960s when he analyzed an apparently obscure phenomenon in bacteria, discovered 10 years earlier by Bertani and Weigle, called host-controlled modification. Arber ...
ChIP-on-chip - Wikipedia, the free encyclopedia
... What is ChIP-on-chip? ChIP-on-chip, also known as genome-wide location analysis, is a technique that is used by scientists in order to investigate Protein-DNA interactions. This technique combines elements from chromatin immunoprecipitation (ChIP) with microarray technology (chip) hence giving it th ...
... What is ChIP-on-chip? ChIP-on-chip, also known as genome-wide location analysis, is a technique that is used by scientists in order to investigate Protein-DNA interactions. This technique combines elements from chromatin immunoprecipitation (ChIP) with microarray technology (chip) hence giving it th ...
II. The selected examples
... Mot box (Fig. 7.9). These promoters required the phageencoded MotA and AsiA proteins, the products of delay-early genes. AsiA protein binds to region 4 ofσ70 and inhibits its to the - 35 sequence. AsiA allows MotA to bind to region 4, it can now recognize the - 30 sequence of the middle T4 promoter. ...
... Mot box (Fig. 7.9). These promoters required the phageencoded MotA and AsiA proteins, the products of delay-early genes. AsiA protein binds to region 4 ofσ70 and inhibits its to the - 35 sequence. AsiA allows MotA to bind to region 4, it can now recognize the - 30 sequence of the middle T4 promoter. ...
9/30 - Utexas
... 2. Gene expression takes time: Typically more than an hour from DNA to protein. Most rapidly 15 minutes. Fig 15.1 ...
... 2. Gene expression takes time: Typically more than an hour from DNA to protein. Most rapidly 15 minutes. Fig 15.1 ...
week9_DNA&geneExpression.bak
... 2. Transcriptional Control - Eukaryotic Gene Expression • OFF: proteins are produced that bind to gene preventing RNA polymerase from binding ...
... 2. Transcriptional Control - Eukaryotic Gene Expression • OFF: proteins are produced that bind to gene preventing RNA polymerase from binding ...
S. cerevisiae Positive Control Primer Set ACT1
... Background: The S. cerevisiae Positive Control Primer Set ACT1 amplifies a 121 base pair fragment from the coding region of the S. cerevesiae ACT1 gene. It can be used as a control for RNA pol II phospho-Ser 2. Contents: This control primer set contains both forward and reverse primers in 400 µl of ...
... Background: The S. cerevisiae Positive Control Primer Set ACT1 amplifies a 121 base pair fragment from the coding region of the S. cerevesiae ACT1 gene. It can be used as a control for RNA pol II phospho-Ser 2. Contents: This control primer set contains both forward and reverse primers in 400 µl of ...
Chp 7 DNA Structure and Gene Function 1
... 2. What is the relationship between a gene and a protein? 3. What are the steps of translation? 4. Where in the cell does translation occur? 5. What are the types of mutations, and how does each alter the encoded protein? ...
... 2. What is the relationship between a gene and a protein? 3. What are the steps of translation? 4. Where in the cell does translation occur? 5. What are the types of mutations, and how does each alter the encoded protein? ...
Chapter 10 Structure and Function of DNA
... Mutations may result from: Errors in DNA replication Physical or chemical agents called mutagens ...
... Mutations may result from: Errors in DNA replication Physical or chemical agents called mutagens ...