* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download Fish Taxonomy and Systematics_Lecture 3
Survey
Document related concepts
Inclusive fitness in humans wikipedia , lookup
The eclipse of Darwinism wikipedia , lookup
Theistic evolution wikipedia , lookup
Paleontology wikipedia , lookup
Evolutionary history of life wikipedia , lookup
Evolving digital ecological networks wikipedia , lookup
Punctuated equilibrium wikipedia , lookup
Coevolution wikipedia , lookup
Evidence of common descent wikipedia , lookup
Sociobiology wikipedia , lookup
Hologenome theory of evolution wikipedia , lookup
Transitional fossil wikipedia , lookup
Saltation (biology) wikipedia , lookup
Transcript
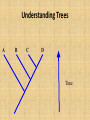
Fish Taxonomy and Systematics Lecture 3: The Evolutionary Relationships Among Populations, Species And Higher Taxa Why Systematics? Organization Basis for identification/ classification of life Gives insight into biological processes: speciation processes adaptation to environment Understanding relationships Common language! Systematics Understand patterns of diversity How? ...in the context of evolutionary and ecological theory. trends in where fish groups are found (spatial distribution) trends in emergence/extinction of evolutionary groups The Concept of “Relationship” Morphological (Linnaeus): the smallest group of individuals that look different from each other. can misclassify based on differences that can be maintained within an interbreeding group depends only on observable morphological differences The Concept of “Relationship” Biological (Mayr): group of populations of individuals that are similar in form and function and that are reproductively isolated from other populations conventional definition until late 1980’s includes genetic information ignores hybridization dependent on geographic isolation to achieve species status The Concept of “Relationship” Evolutionary (Wiley): a population or group of populations that shares a common evolutionary fate and historical tendencies recognizes more than just genetic and morphological differences difficult to determine “evolutionary fate” how much diversity is allowed within a common evolutionary fate? Nelson 1999 Reviews in Fish Biology and Fisheries The Concept of “Relationship” Phylogenetic: the smallest biological unit appropriate for phylogenetic analysis (process that rates traits as ancestral (plesiomorphes) or derived (apomorphies) and then looks for groupings based on similarities (shared, synapomorphies) does not infer modes of speciation nothing is arbitrary depends on thorough phylogenetic analysis first The Concept of “Relationship” Usefulness of each concept depends on the use - for Endangered Species Act, use as much evidence as possible: morphological, physiological, behavioral geographic life history & development habitat & feeding ecology phylogenetics evolutionary fate Determining Relationships Between Taxa Traditional: examine and list primitive to advanced, link groups based on a few arbitrary traits, generate lineage model based on these limited data Determining Relationships Between Taxa Phenetics: multivariate statistical approach: assemble list of traits determine degree of similarity among groups based on number of similar traits operates on the assumption that the total phenotype accurately reflects the genotype. has been largely a failure when applied to higher organisms (Ernst Mayr -Evolution and the Diversity of Life, 1976, p. 429) ignores evolutionary linkage of groups (convergence could put evolutionarily distinct lines into a single taxon) Determining Relationships Between Taxa Evolutionary approach: for the evolutionary systematist, relationship means more than just kinship in a strictly genealogical sense it also involves a measure of genetic change that may occur within a group subsequent to its divergence (branching) from an ancestral group. Determining Relationships Between Taxa Phylogenetic (cladistic): define relationship in a strictly genealogical sense—the measure of phylogenetic relationship is the relative recency of common descent assemble a list of traits classify each taxonomic group on basis of presence or absence of each trait determine degree of similarity among groups based on shared and unique traits: Determining Relationships Between Taxa Phylogenetic (cladistic), continued: determine degree of similarity among groups based on shared and unique traits: 1. shared traits = plesiomorphic traits (ancestral) 2. unique traits = apomorphic traits (derived) 3. shared unique traits = synapomorphic traits monophyletic group of taxa (common origin) = clade Advantage: – Classification reflects pattern of evolution – Classification not ambiguous Imagine an ancestral species A that gives rise to three modern-day species, B, C, and D. Imagine further that 15% of the genetic content of species B differs from that of species A, 10% of the genetic content of species C differs from that of species A, and 70% of the genetic content of species D differs from that of species A. Evolutionary approach There is a maximum genetic difference of 25% between the genomes of B and C, but 80% between C and D. The evolutionist would say B and C are more closely related than either is to D, because there is much less genetic difference or genetic change between them, that is, they have a much greater inferred amount of shared genotype. Cladistic approach The cladist, in direct opposition to the evolutionary systematist, would conclude that C is more closely related to D than to B because of greater recency of common descent. Over the years, the cladistic approach has become widely accepted over any alternative approach, so that today nearly everyone in the field of biosystematics is a cladist! Cladistic Systematics Some recent classifications attempt to show only evolutionary relationships among organisms, ignoring their degree of morphological similarity or difference. The objective of cladistic systematics is to determine the evolutionary histories of organisms and then to express those relationships in phylogenetic trees. A clade is the entire portion of a phylogeny that is descended from a single ancestral species. The closeness of organisms on a cladogram indicates the presumed time since they diverged from their most recent common ancestor. Because the goal is to show phylogenies, taxa in a cladistic classification are clades and are monophyletic, i.e., each taxon is a single lineage that includes all-and only-the descendants of a single ancestor. Traits shared due to descent from a common ancestor are called ancestral traits. Q. How can ancestral traits be recognized? A good fossil record helps reveal ancestral traits. For example, the excellent fossil record of horses shows that modern horses, which have one toe on each foot, evolved from ancestors that had multiple toes. A trait, such as the modern horse's single toe, that differs from the ancestral trait in the lineage is called a derived trait. To erect classification systems that accurately reflect phylogenies, it is necessary to be able to distinguish ancestral from derived traits. Therefore, cladists devote much effort to gathering and interpreting evidence to determine which traits are really ancestral and which are derived in different groups of organisms. To erect classification systems that accurately reflect phylogenies, it is necessary to be able to distinguish ancestral from derived traits. Therefore, cladists devote much effort to gathering and interpreting evidence to determine which traits are really ancestral and which are derived in different groups of organisms. Study of Mitochondrial and Chloroplast genomes DNA in these two organelles is useful in evolutionary studies because: Genomes have a relatively small size, especially when compared to nuclear DNA They are well organized, discrete units of inheritance There are few recombination events Uniparental inheritance (usually) Often high copy-numbers in the cell (i.e. easy to work with) Mutation rates vary (useful to study many levels of diversity) What is a phylogeny? A phylogeny is a type of pedigree Shows relationships between species, not individuals Reconstructs pattern of events leading to the distribution and diversity of life Often shown as a network or tree Understanding Trees A B C D Time Understanding Trees A B C D “Root”: common ancestor of organisms in the phylogeny Understanding Trees A B C D Internal branch: common ancestor of a subset of species in the tree Understanding Trees A B C D “Node”: point of divergence of two species Understanding Trees A B C D “Leaf”: terminal branch leading to a species Understanding Trees A B C D Clade: group of species descended from a common ancestor Why study phylogenies? It is useful to know how organisms are related Taxonomy Character evolution and state prediction Ecology Co-evolution Biogeography Divergence times Medicine Uses of phylogenies: Taxonomy Similar organisms are grouped together Clades share common evolutionary history Phylogenetic classification names clades Source: Pryer, K.M., H. Schneider, A.R. Smith, R. Cranfill, P.G. Wolf, J.S. Hunt and S.D. Sipes. 2001. Horsetails and ferns are a monophyletic group and the closest living relatives to seed plants. Nature 409: 618622 Source: Inoue, J.G., Miya, M., Tsukamoto, K., Nishida, M. 2003. Basal actinopterygian relationships: a mitogenomic perspective on the phylogeny of the “ancient fish”. Molecular Phylogenetics and Evolution, 26: 110-120. Uses of phylogenies: Character evolution Examine changes in particular traits e.g. body plan in animals Predict similar traits in related species e.g. Taxol (a cancer drug) in the Yew tree Correlation between 2 characters e.g. fruit traits and dispersal mechanism in plants Example of correlated character evolution After: Conner, J.K. 2002. Genetic mechanisms of floral trait correlations in a natural population. Nature 420: 407-410.Conner, J.K. 1997. Floral Evolution in Wild Radish: The Roles of Pollinators, Natural Selection, and Genetic Correlations Among Traits. Int. J. Plant Sci. 158(6 Suppl.): S108-S120.Conner, J. and S. Via. 1993. Patterns of phenotypic and genetic correlations among morphological and life history traits in wild radish, Raphanus raphanistrum. Evolution 47: 704-711. Corolla and Filament length in Flowers Uses of phylogenies: Ecology Study the evolution of ecological interaction and behavior Why might two related species have a different ecology? e.g. social vs. solitary, drought tolerant vs. mesophytic, parasitic vs. free living, etc. What are the causes of these differences? Is the environment causing these differences? Can we infer which condition is ancestral? Examples of phylogenetic ecology Evolutionary ecology of mate choice in swordtail fish (genus Xiphophorus) M. R. MORRIS, P. F. NICOLETTO & E. HESSELMAN. 2003. A polymorphism in female preference for a polymorphic male trait in the swordtail fish Xiphophorus cortezi. ANIMAL BEHAVIOUR, 65, 45–52 doi:10.1006/anbe.2002.2042. G. G. Rosenthal, T. Y. Flores Martinez, F. J. Garcîa de Leo, and M. J. Ryan. 2001. The American Naturalist. vol. 158, no. 2 Shared Preferences by Predators and Females for Male Ornaments in Swordtails.G. G. ROSENTHAL AND C. S. EVANS. 1998. Evolution Female preference for swords in Xiphophorus helleri reflects a bias for large apparent size (sexual selection Uses of phylogenies: Co-evolution Compare divergence patterns in two groups of tightly linked organisms (e.g. hosts and parasites or plants and obligate pollinators) Look at how similar the two phylogenies are Look at host switching Evolutionary arms races Traits in one group track traits in another group e.g. toxin production and resistance in prey/predator or plant/herbivore systems, floral tube and proboscis length in pollination systems Example of host-parasite phylogeny Source: Page, R.D.M., Cruickshank, R.H., Dickens, M., Furness, R.W., Kennedy, M., Palma, R.L., Smith, V.S. 2004. Phylogeny of “Philoceanus complex” seabird lice (Phthiraptera: Ischnocera) inferred from mitochondrial DNA sequences. Molecular Phylogenetics and Evolution, 30: 633-652. Uses of phylogenies: Phylogenetic geography Sometimes called historical biogeography or phylogeography Map the phylogeny with geographical ranges of populations or species Understand geographic origin and spread of species e.g. origin of modern humans in Africa Look at similarities organisms between unrelated Understand repeated patterns in distributions e.g. identifying glacial refugia Uses of phylogenies: Estimating Divergence Times Estimate when a group of organisms originated Uses information about phylogeny and rates of evolutionary change to place timescales on tree Needs calibration with fossils Combined with mapping characters, correlate historical events with character evolution e.g. Radiation of flowering plants in the Cretaceous Uses of phylogenies: Medicine Learn about the origin of diseases Look for disease resistance mechanisms in other hosts to identify treatment and therapy in humans Multiple origins of HIV from SIV (Simian Immunodeficiency Virus) From: Understanding Evolution. HIV: the ultimate evolver. http://evolution.berkeley.edu/evolibrary/article/0_0_0/medicine_04 Example of disease phylogeny Sourc: Understanding Evolution: Tracking SARS back to it's source. http://evolution.berkeley.edu/evolibrary/news/060101_batsars After: Wendong Li, et al. 2005. Bats Are Natural Reservoirs of SARS-Like Coronaviruses. Science 28 October 2005: Vol. 310. no. 5748, pp. 676 - 679. DOI: 10.1126/science.1118391 Phylogeny in medical forensics: HIV A dentist who was infected with HIV was suspected of infecting some of his patients in the course of treatment HIV evolves very quickly (10-3 substitutions/year) Possible to trace the history of infections among individuals by conducting a phylogenetic analysis of HIV sequences Samples were taken from dentist, patients, and other infected individuals in the community Study found 5 patients had been infected by the dentist Source: Ou et. al. 1992. Molecular epidemiology of HIV transmission in a dental practice. Science, 256: 1165-1171.