* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download What is virulence
Triclocarban wikipedia , lookup
Quorum sensing wikipedia , lookup
Marine microorganism wikipedia , lookup
Neonatal infection wikipedia , lookup
Horizontal gene transfer wikipedia , lookup
Bacterial cell structure wikipedia , lookup
Lyme disease microbiology wikipedia , lookup
Globalization and disease wikipedia , lookup
Gastroenteritis wikipedia , lookup
Sarcocystis wikipedia , lookup
Schistosoma mansoni wikipedia , lookup
Hospital-acquired infection wikipedia , lookup
African trypanosomiasis wikipedia , lookup
Sociality and disease transmission wikipedia , lookup
Transmission (medicine) wikipedia , lookup
Molecular mimicry wikipedia , lookup
Bacterial morphological plasticity wikipedia , lookup
Infection control wikipedia , lookup
Human microbiota wikipedia , lookup
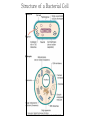
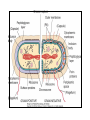
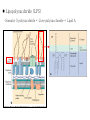
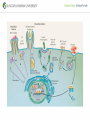
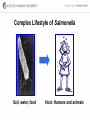
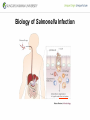
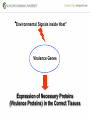
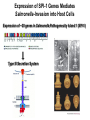
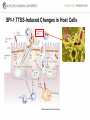
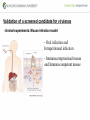
Genetic Screenings for Studying Bacterial Pathogenesis Dongwoo Shin, Ph.D. Associate Professor, Department of Molecular Cell Biology, Sungkyunkwan University School of Medicine Structure of a Bacterial Cell What is a pathogen? -An organism capable of colonizing a host organism where the interaction results in disease -opportunistic pathogens -strict pathogens Opportunistic pathogens - Most of infections - Normal microbiota for causing disease - No disease in normal setting but disease when introduced into unprotected sites (e.g. blood, tissues) - Immunocompromised patients are more susceptible Staphylococcus aureus Escherichia coli Strict pathogens - A few infections Mycobacterim tuberculosis (tuberculosis) Neisseria gonorrhoeae (gonorrhea) [NOTE] Colonization vs. Infection Exposure of an individual to an organism 1. Transient (hours or days) colonization 2. Permanent colonization 3. Disease production (Infection) [NOTE] Normal microbiota (microflora) In a healthy human, - The internal tissues (e.g. brain, blood, muscles): normally free of microorganisms - Conversely, the surface tissues (e.g. skin and mucous membrane): constantly in contact with environmental microorganisms and become colonized by certain microbial species - Normal microbiota (= microflora, normal flora): mixture of microorganisms regularly found at any anatomical site - Bacteria make up most of the normal microbiota over the fungi and protozoa What is virulence (pathogenicity)? -The capacity of a pathogen to cause damage or disease in the host -Virulence factors: Cell wall components (LPS, LTA), DNA, Proteins Lipopolysacchride (LPS) -Somatic O polysacchride + Core polysaccharide + Lipid A What is virulence (pathogenicity)? -The capacity of a pathogen to cause damage or disease in the host -Virulence factors: Cell wall components, DNA, Proteins Bacteria and Disease Establishing connection: Koch’s Postulates • Proving cause and effect in infectious disease research • First raised in the 1800s by Robert Koch • Koch’s postulates -association of the bacteria with the lesions of the disease -isolating the bacterium in pure culture -showing that the isolated bacterium causes disease in humans or animals -reisolating the bacterium from the intentionally infected animal Bacteria and Disease Establishing connection: Molecular Koch’s Postulates • Raised in the 1988 by Stanley Falkow • Molecular Koch’s postulates -gene (or its product) should be found only in strains of bacteria that cause the disease -gene should be “isolated” by cloning -disruption of gene in virulent strain should reduce virulence -gene is expressed by bacterium during infectious process in animal or human Identification of bacterial virulence factors 1. Understanding the molecular strategies used by a pathogen during host infection 2. Providing the targets in development of novel therapeutics for bacterial infection -Currently used antibiotics → Targeting bacterial viability → Selective pressure -Antivirulence therapy Salmonella Enterobacteriaceae Escherichia coli Shigella Serovars Salmonella enterica Salmonella bongori Typhimurium Enteritidis Typhi Paratyphi Gastroenteritis Systemic disease Complex Lifestyle of Salmonella Soil, water, food Host: Humans and animals Salmonella enterica serovar Typhimurium A facultative intracellular pathogen Infects millions of people worldwide every year resulting in ~500,000 deaths Transmission via contaminated food or water Gastroenteritis Serves (Human) and Typhoid fever (Mouse) as a model system for other intracellular pathogens Biology of Salmonella Infection “Environmental Signals inside Host” Virulence Genes Expression of Necessary Proteins (Virulence Proteins) in the Correct Tissues Stomach Invasion into epithelial cells of small intestine Type III Secretion Systems (TTSSs) Survival inside macrophages Systemic disease Expression of SPI-1 Genes Mediates Salmonella-Invasion into Host Cells Expression of ~30 genes in Salmonella Pathogenecity Island 1 (SPI-1) Type III Secretion System SPI-1 TTSS-Induced Changes in Host Cells Stomach Invasion into epithelial cells of small intestine Type III Secretion Systems (TTSSs) Survival inside macrophages Systemic disease Bacterial killing processes inside phagosome Antimicrobial peptides Phagosome Bacterial Killing ROS & RNS production Phagosome-lysosome fusion Induction of the SPI-2 T3SS within Macrophage Host cells effector proteins secretion system Salmonella The SPI-2 T3SS prevents bacterial killing by macrophages Antimicrobial peptides LPS modification Phagosome SPI-2 TTSS Preventing recruitment of NADH oxidase ROS & RNS production Alteration of vesicle trafficking Phagosome-lysosome fusion Genetic screenings of bacterial virulence factors 1. Pre-genomic era: 1990’s 2. Post-genomic era: 21C In Vivo Expression Technology (IVET) 1. Isolation of Salmonella genes whose expression is induced inside the host (i.e. genes whose products are necessary for host infection) 2. Auxotrophic selection method (Science, 1993) and Differential fluorescence induction method (Science, 1997) Auxotrophic selection method Step 1: Creating transcriptional fusions of random fragments of the Salmonella chromosome with promoterless purA and lacZ genes; introduction of this library into a purA mutant a mutant that cannot synthesize purines (auxotroph) Auxotrophic selection method Step 2: Integration of a plasmid construct into chromosome of a purA mutant via single crossover Auxotrophic selection method Step 3: Host infection with the pool of fusion strains and selection X-gal plate Differential fluorescence induction (DFI) Rationale: trapping the gene promoters that are activated inside macrophages; using GFP PmgtC gfp Activation of mgtC transcription Salmonella macrophage Differential fluorescence induction (DFI) Step 1: Cloning of random fragments of Salmonella chromosome into a promoterless gfp plasmid; introduction of plasmids into Salmonella Differential fluorescence induction (DFI) Step 2: Infection of macrophages with Salmonella harboring gfp fusion plasmids; sorting GFP-active Salmonella with FACS Validation of a screened candidate for virulence 1. In vitro method: Gentamicin protection assay for evaluations of Salmonella invasion into and survival within host cells 2. In vivo method: Animal experiments for evaluations of Salmonella’s ability to infect host Validation of a screened candidate for virulence - Gentamicin protection assay: Infection of epithelial cells (e.g. Hep-2) or macrophages (e.g. J774.A) with Salmonella Incubation allowing for Salmonella to invade into epithelial cells or for macrophages to engulf Salmonella (i.e. phagocytosis) Gentamicin treatment to kill bacteria outside host cells Detergent treatment to lyse host cells; plating onto agar plate to count Salmonella Validation of a screened candidate for virulence - Gentamicin protection assay: a mutant that cannot produce SPI-2 TTSS Validation of a screened candidate for virulence - Animal experiments: Mouse infection model - Oral infection and Intraperitoneal infection - Immunocompromised mouse and Immunocompetent mouse Validation of a screened candidate for virulence - Animal experiments: Genetic screenings of bacterial virulence factors 1. Pre-genomic era: 1990’s 2. Post-genomic era: 21C Announcement of Salmonella genome sequence: Nature (2000) “Now, one can predict which genes are important for Salmonella virulence and experimentally test them.” In this paper, the authors evaluated the role of every single transcription factor (83 regulators) in Salmonella virulence - A revolutionary method for construction gene deletion mutants in E. coli: PNAS (2000) - Applicable to other enteric bacteria: Salmonella, Klebsiella, Yersinia, Enterobacter etc. - Accurate, fast, and cheap method Identification of 14 regulators required for Salmonella virulence