* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download Evolution, 2e
Survey
Document related concepts
DNA barcoding wikipedia , lookup
Extrachromosomal DNA wikipedia , lookup
History of genetic engineering wikipedia , lookup
Therapeutic gene modulation wikipedia , lookup
Nucleic acid analogue wikipedia , lookup
Deoxyribozyme wikipedia , lookup
Helitron (biology) wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Quantitative comparative linguistics wikipedia , lookup
Metagenomics wikipedia , lookup
Microevolution wikipedia , lookup
Viral phylodynamics wikipedia , lookup
Transcript
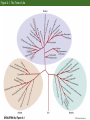
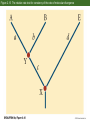
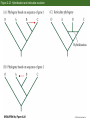
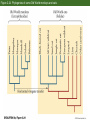
Classification and Phylogenies • Taxonomic categories and taxa • Inferring phylogenies – – – – – – The similarity vs. shared derived character states Homoplasy Maximum parsimony, maximum likelihood, and Baysian methods An example of phylogenetic analysis Molecular clocks and timing of branching events Difficulties in Phylogenetic Analysis • Taxonomic categories – – – – – – – Species Genus Family Order Class Phylum Division Chapter 2 Opener How do we classify organisms? Morphological similarity occassionally obscurs relationships Figure 2.2 Darwin’s representation of hypothetical phylogenetic relationships Figure 2.1 The Tree of Life Figure 2.5 An example of phylogenetic analysis applied to three data sets (Part 1) Sometimes, phylogenies derived from similarities are congruent with a phylogeny derived from synapomorphies Figure 2.5 An example of phylogenetic analysis applied to three data sets (Part 2) Similarities can produce an incorrect phylogeny Figure 2.5 An example of phylogenetic analysis applied to three data sets (Part 3) Homoplasies confound phylogeny reconstruction Figure 2.6 Monophyletic groups whose members share derived character states that evolved only once Easy reconstruction Figure 2.7 Two possible hypotheses for the phylogenetic relationships of humans Principle of parsimony: Okkam’s razor Figure 2.9 Members of the primate superfamily Hominoidea Phenetic vs. cladistic classifications Figure 2.10 Evidence for phylogenetic relationships among primates, based on the ψη-globin pseudogene mtDNA 4,700 base sequence Genes for 11 tRNAs 6 proteins Human-chimpanzee relationship 1023 more likely than Chimpanzee-gorilla relationship Y DNA Base sequence for Testis-specific protein Y Autosomal DNA Base sequence of Beta-globin gen cluster Figure 2.11 Relationships among major groups of vertebrates Morphological and DNA sequences sometimes reveal the same phylogeny Figure 2.13 (A) If divergence occurred at a nearly constant rate, relative times of divergence of lineages could be determined from differences/similarities between taxa and phylogeny of the taxa could then be estimated. (B) Hypothetical phylogeny in which evolution occurs at a nearly constant rate Figure 2.14 Calibration of molecular clocks in Hawaiian organisms Slope of regression reveals the rate of evolution Y = a + bX b = 0.016 b = 0.019 Fruit fly divergence: Hawaiian Islands Results of speciation Figure 2.15 The relative rate test for constancy of the rate of molecular divergence Difficulties in phylogenetic reconstruction • 1. Scoring characters • 2. Homoplasy • 3. Past evolutionary events may be obscured by recent evolution. • 4. Polytomy • 5. Gene trees and character trees can be incongruent • 6. ± hybridization and horizontal allele transfer Figure 2.23 Hybridization and reticulate evolution Figure 2.24 Phylogenies of some Old World monkeys and cats