* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download Functional Genomics
Survey
Document related concepts
List of types of proteins wikipedia , lookup
Transcriptional regulation wikipedia , lookup
Gene expression wikipedia , lookup
Non-coding DNA wikipedia , lookup
Community fingerprinting wikipedia , lookup
Genomic imprinting wikipedia , lookup
Gene desert wikipedia , lookup
Promoter (genetics) wikipedia , lookup
Ridge (biology) wikipedia , lookup
Molecular evolution wikipedia , lookup
Silencer (genetics) wikipedia , lookup
Gene regulatory network wikipedia , lookup
Endogenous retrovirus wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Transcript
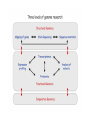
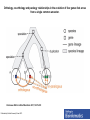
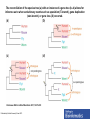
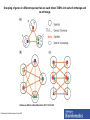
Functional Genomics Branches of Genomics Structural Genomics Metagenomics Genomics Comparativ e Genomics Functional Genomics Why We Need Functional Genomics Organism E. coli yeast C. elegans Drosophila Arabadopsis mouse human # genes % of genes with inferred function Completion date of genome 4288 6,600 19,000 12-14K 25,000 ~30,000? ~30,000? 60 40 40 25 40 10-20 10-20 1997 1996 1998 1999 2000 2002 2000 How to determine functionally related genes? • 40% if predicted genes in newly sequenced genomes cannot be assigned function based on sequence similarity. • Genes sharing a common pattern of expression in many different experiments are likely to be involved in similar processes. – Gene A regulates Gene B, or vice versa – Gene A and Gene B are regulated by Gene C Orthology, co-orthology and paralogy relationships in the evolution of four genes that arose from a single common ancestor. Kristensen D M et al. Brief Bioinform 2011;12:379-391 Published by Oxford University Press 2011. The reconciliation of the species tree (a) with an instance of a gene tree (b–d) allows for inference as to when evolutionary events such as speciation (T-branch), gene duplication (star-branch), or gene loss (X) occurred. Kristensen D M et al. Brief Bioinform 2011;12:379-391 Published by Oxford University Press 2011. Grouping of genes in different species that are each others’ BBHs into sets of orthologs and co-orthologs. Kristensen D M et al. Brief Bioinform 2011;12:379-391 Published by Oxford University Press 2011. Clusters of Orthologous Groups (COG) • Identification of orthologs is critical for reliable prediction of gene function in newly sequenced genomes. • The purpose of COG is to serve as a platform for functional annotation of newly sequenced genomes and for study of genome evolution. Links • Unicellular clusters (COGs): – http://www.ncbi.nlm.nih.gov/COG/grace/uni.html • Eukaryotic orthologous groups (KOGs): – http://www.ncbi.nlm.nih.gov/COG/grace/shokog.cgi • 2003 paper • 2011 paper