* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download Janeway`s Immunology - Cal State LA
Survey
Document related concepts
Monoclonal antibody wikipedia , lookup
Cancer immunotherapy wikipedia , lookup
Duffy antigen system wikipedia , lookup
Antimicrobial peptides wikipedia , lookup
DNA vaccination wikipedia , lookup
Immune system wikipedia , lookup
Innate immune system wikipedia , lookup
Adoptive cell transfer wikipedia , lookup
Adaptive immune system wikipedia , lookup
Human leukocyte antigen wikipedia , lookup
Polyclonal B cell response wikipedia , lookup
Transcript
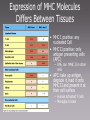
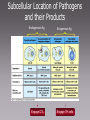
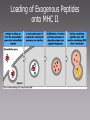
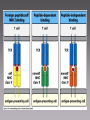
MICR 304 Immunology & Serology Lecture 9 TCR, MHC molecules Chapter 3.10 – 3.19, 4.9- 4.11, 5.1 – 5.19 Overview of Today’s Lecture • Generation of T cell receptor (TCR) • MHC molecules • Antigen presentation via MHC molecules Key Players in Immunology Innate Adaptive Cells Phagocytes Epithelial Cells NK Cells Lymphocytes (B-Ly, T-Ly) Effector molecules Complement Antimicrobial (Poly)Peptides Antimicrobial Lipids? Antibodies T Cell Receptor • 2 chains – Connected by disulfide bond – Variable region – Constant region – Short cytoplasmic tail • Mostly a and b chain • Some specialized Tcells have g and d chain (gd T cells) T-Cell Receptor Belongs to the Immunoglobulin Super Family Gene Segments Coding for the T-Cell Receptor • Variable region of a-chain is composed of V and J gene segments • Variable region of b-chain is composed of V, D and J gene segments Generation of the TCR by Gene Rearrangement and Recombination No secondary modification of TCRs Diversity of the Lymphocyte Antigen Receptors P- and N- nucleotides: nucleotides added during initial gene rearrangement and recombination Unique Properties of TCR Compared to BCR • Only one antigen binding site • Never secreted • Recognize processed antigen presented through specialized molecules TCR Recognizes Antigen Presented by MHC Molecules • MHC: major histocompatibility complex • First identified in transplantation immunology • T cells recognize antigen bound to an MHC molecule • Two types of MHC molecules – MHC I: presents endogenous peptides MHC I CTL – MHC II: presents exogenous peptides MHC II TH • Virus encoded • Produced by intracellularly replicating microorganisms • Tumor antigens • Uptake through phagocytosis and degradation in phagolysosome Antigen Recognition through TCR Requires Additional Molecules • CD4: Interaction with MHC II • CD8: Interaction with MHC I TH • CD3: signal transduction CTL Cytokines Ag presenting cell Any nucleated cell Cytotoxic granules Contrast TH Cells and CTL TH CTL CD on surface CD4 CD8 MHC Interaction Target cells MHC II MHC I APC (macrophages, DC, B cells, others) Cytokine release Any nucleated cell Response upon activation Cytotoxic granule release (and some cytokines) gd T cells are Distinct • Minority of T cell population • Bind heat shock proteins and nonpeptide ligands – Mycobacterial lipid antigen – Phosphorylated ligands • Not restricted by classical MHC I or MHC II molecules • May bind to free antigen Expression of MHC Molecules Differs Between Tissues • MHC I positive: any nucleated Cell • MHC II positive: only antigen presenting cells (APC) – IFNg can MHC II in other cells • APC: take up antigen, degrade it, load it onto MHC II and present it at their cell surface – Human activated T cells – Microglia in brain Structure of MHC Molecules MHC I • Endogenous peptides are bound to MHC I by their ends – Ionic interaction • 8 – 10 aa MHC II • Exogenous peptides are bound to MHC II along the groove • 13 - 17 aa Intracellular Compartment Exogenous peptides Endogenous compartment Subcellular Location of Pathogens and their Products Endogenous Ag Engage CTL Exogenous Ag Engage TH cells Key Molecules in Antigen Presentation MHC I MHC II • Proteasomes • TAP1, 2 (Transporters associated with antigen processing) • CD8 • Lysosomal proteases • CLIP (class II associated invariant chain peptide) • B7 • CD4 • CD28 On target cell On CTL On APC On TH Loading of Endogenous Peptides onto MHC I (1) • Chaperones guide in ER nascent MHC I to TAP • Endogenous proteins are degraded in proteasome and enter ER through TAP • Peptide loading occurs in the ER • If peptides are too long they can be trimmed the endoplasmic reticulum aminopeptidase associated with antigen processing (ERAAP) • MHC I with peptide loaded is sent to cell surface Loading of Endogenous Peptides onto MHC I (2) Viral Strategies to Interrupt Active LearningofExercise: Presentation Viral Peptides • Blocking entry of viral peptides into ER By which mechanisms could • Retention of MHC I in ER • Degradation of MHCwith I via transport of viruses interfere the MHC I into cytosol presentation of viral peptides on • Blocking access of CTLs to surface MHC I at the cellloaded surface? expressed peptide MHC I Loading of Exogenous Peptides onto MHC II Loading of Exogenous Peptides onto MHC II (1) • MHC II leaves ER with CLIP – CLIP • Class II associated invariant-chain peptide • Binds to peptide groove • Prevents premature peptide loading • MHC II vesicle fuses with phagosome containng degraded exogenous peptides • HLA-DM removes CLIP from MHC II peptide groove and exogenous degraded peptide can bind – HLA-DM is MHC II like Loading of Exogenous Peptides onto MHC II (2) Limitations in MHC Binding Pose a Problem • How can so many different pathogen derived peptides be presented? •Introduce variability in MHC molecule •MHC is polymorphic •MHC is polygenic Polymorphism and Polygeny Increase Variability • Polymorphism – Numerous variants (alleles) for each gene – MHC genes are the most polymorph genes known • Polygeny – Several different genes for MHC I and MHC II – A set of genes with a broader range of peptide binding is expressed Genes Coding for MHC Molecules Polygeny • 3 genes and gene products for MHC I – A (a) – B (a) – C (a) – b2 microglobulin is monomorphic • 3 genes and 4 gene products for MHC II – DR (a, b1, b2) – DP (a,b) – DQ (a,b) Polymorphism • Over 1000 alleles • Alleles are codominant expressed Number of Different Alleles Polymorphism of MHC Genes … a growing list! Allelic Variation Occurs at Specific Sites with in the MHC Molecules The Expression of MHC Alleles is Co-Dominant Intra- and Interpersonal Variability of MHC Molecules • Within a person multiple MHC molecules are expressed – 3 MHC I genes x 2 (father, mother) = 6 MHC I – 4 sets of MHC II genes x 2 (father, mother) = 8 MHC II • Cells within a person are uniform • Cells from another persons carry different sets of MHC molecules! T Cell Recognition of Antigens is MHC Restricted MHC molecules participate in antigen recognition. Superantigens • Antigens that are not processed • Crosslink TCR with MHC • Can simultaneously stimulate 2 - 20% of all T cells. Non-Classical MHC Genes • • • • • Resemble MHC class I genes in structure Many associate with b2microglobulin Also called MHC Ib Comparatively little polymorphism Some bind to activating NK cell receptors (NKG2D) – Example: MIC-A – Induced in response to cellular stress – Trigger cytotoxicity • Some bind to NK inhibitory receptors (NKG2A) – Example: HLA E – Inhibit cytotoxicity – Found on fetus derived placental cells • Some present lipid antigen to T cells – Example CD1 Refresher: NK Cell Mediated Killing Today’s Take Home Message • The TCR consists of two chains, a and b, and is similar to the arm of an antibody molecule with the TCR a-chain representing the light chain and the b-chain the heavy chain. • T cell recognize digested peptide presented through MHC molecules. • T helper cells recognize peptide on MHC II and utilize CD4 to ensure proper binding to MHC II. • CTL recognize peptide on MHC I and utilize CD8 to ensure proper binding to MHC I. • MHC are polymorphic and polygenic. • Non-classical MHC I molecules (MHC Ib) interact with inhibitory and activating NK cell receptors.