* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download Regulation of yeast mating - City University of New York
Non-coding DNA wikipedia , lookup
Genetic engineering wikipedia , lookup
Epigenetics in stem-cell differentiation wikipedia , lookup
Point mutation wikipedia , lookup
Epigenetics in learning and memory wikipedia , lookup
Genome evolution wikipedia , lookup
Ridge (biology) wikipedia , lookup
Gene expression programming wikipedia , lookup
Microevolution wikipedia , lookup
Biology and consumer behaviour wikipedia , lookup
Epigenetics of diabetes Type 2 wikipedia , lookup
Gene therapy of the human retina wikipedia , lookup
Genomic imprinting wikipedia , lookup
History of genetic engineering wikipedia , lookup
Epigenetics of neurodegenerative diseases wikipedia , lookup
Minimal genome wikipedia , lookup
Genome (book) wikipedia , lookup
Long non-coding RNA wikipedia , lookup
Nutriepigenomics wikipedia , lookup
Site-specific recombinase technology wikipedia , lookup
Vectors in gene therapy wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Gene expression profiling wikipedia , lookup
Polycomb Group Proteins and Cancer wikipedia , lookup
Designer baby wikipedia , lookup
Primary transcript wikipedia , lookup
Epigenetics of human development wikipedia , lookup
Transcription factor wikipedia , lookup
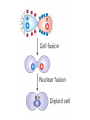
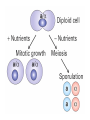
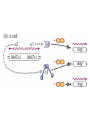
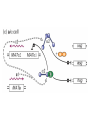
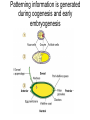
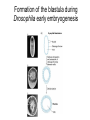
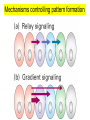
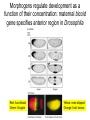
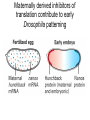
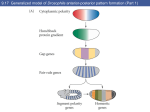
Lecture 10 Gene Control in Development Cell type specification Development of an organism Reading: Chapter 11:471-2 Chapter 15.1; 15.3; 15.4 Chapter 22.2 Molecular Biology syllabus web site Cell type specification in the yeast model system Different mating types express specific transcription factors that form complexes with MCM1 MCM1 is a general transcription factor found in all cell types Transcription factors and lessons from yeast • Factors may act alone or in combinations • The same factor in different combinations may contribute to complexes that act as repressors or activators. Cell type specification in animals Cell type specification in mammals: skeletal myogenesis proceeds through three stages How to identify factors involved in muscle development? To identify transcription factors that may play a role in “determination” of cells destined for a specific organ: • Isolation of cDNAs by subtractive hybridization (fibroblasts vs. myoblasts) • Testing by transformation of undetermined cell types to demonstrate effect on “determination” • Create “Knockouts” to confirm information on the stage at which a specific factor acts • Characterization: function as heterodimers (key to specificity is the interaction with other factors) and belong to family of basic helix-loop-helix DNA binding transcription factors (bHLH) MRFs, muscle regulatory factors binding to “E” box in many genes Microarray analysis shows global patterns of gene expression during differentiation Development of an organism: Drosophila melanogaster Drosophila has two life forms Patterning information is generated during oogenesis and early embryogenesis Formation of the blastula during Drosophila early embryogenesis Four maternal gene systems (anterior, posterior, terminal, dorsoventral) control early patterning in fly embryos Mechanisms controlling pattern formation Morphogens regulate development as a function of their concentration: maternal bicoid gene specifies anterior region in Drosophila Red: hunchback Green: Krupple Yellow: even-skipped Orange: fushi tarazu Maternally derived inhibitors of translation contribute to early Drosophila patterning Nanos regulates the translation of Hunchback and helps to establish the Hunchback gradient Use of mutants to characterize Nanos as a translational inhibitor HOX genes and transcription factors discovered through “homeotic mutants” showing transformation of one body part into another Expression domains of Hox genes in Drosophila and mouse embryos Specification of floral-organ identity in Arabidopsis: flowers contain four different organs Three classes of genes control floralorgan identity Expression patterns of floral organidentity genes