* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download Gene Set Enrichment Analysis
Gene therapy of the human retina wikipedia , lookup
Short interspersed nuclear elements (SINEs) wikipedia , lookup
Neuronal ceroid lipofuscinosis wikipedia , lookup
Transposable element wikipedia , lookup
Copy-number variation wikipedia , lookup
Genetic engineering wikipedia , lookup
Epigenetics in learning and memory wikipedia , lookup
Oncogenomics wikipedia , lookup
Long non-coding RNA wikipedia , lookup
Vectors in gene therapy wikipedia , lookup
Gene therapy wikipedia , lookup
X-inactivation wikipedia , lookup
Epigenetics of diabetes Type 2 wikipedia , lookup
Pathogenomics wikipedia , lookup
Quantitative trait locus wikipedia , lookup
Epigenetics of neurodegenerative diseases wikipedia , lookup
Polycomb Group Proteins and Cancer wikipedia , lookup
Gene nomenclature wikipedia , lookup
Essential gene wikipedia , lookup
Public health genomics wikipedia , lookup
History of genetic engineering wikipedia , lookup
Gene desert wikipedia , lookup
The Selfish Gene wikipedia , lookup
Therapeutic gene modulation wikipedia , lookup
Site-specific recombinase technology wikipedia , lookup
Nutriepigenomics wikipedia , lookup
Genome evolution wikipedia , lookup
Genomic imprinting wikipedia , lookup
Minimal genome wikipedia , lookup
Gene expression programming wikipedia , lookup
Ridge (biology) wikipedia , lookup
Biology and consumer behaviour wikipedia , lookup
Microevolution wikipedia , lookup
Epigenetics of human development wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Genome (book) wikipedia , lookup
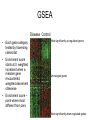
Gene Set Enrichment Analysis GSEA: Key Features • Ranks all genes on array based on their differential expression • Identifies gene sets whose member genes are clustered either towards top or bottom of the ranked list (i.e. up- or down regulated) • Enrichment score calculated for each category • Permutation test to identify significantly enriched categories • Extensive gene sets provided via MolSig DB – GO, chromosome location, KEGG pathways, transcription factor or microRNA target genes GSEA Disease Control • Each gene category tested by traversing ranked list • Enrichment score starts at 0, weighted increment when a member gene encountered, weighted decrement otherwise Most significantly up-regulated genes Unchanged genes • Enrichment score – point where most different from zero Most significantly down-regulated genes Enrichment Score • Enrichment Score (ES) is calculated by evaluating the fractions of genes in S (‘‘hits’’) weighted by their correlation and the fractions of genes not in S (‘‘misses’’) present up to a given position i in the ranked gene list, L, where N genes are ordered according to the correlation, 4 GSEA algorithm GSEA: Permutation Test • Randomise data (groups), rank genes again and repeat test 1000 times • Null distribution of 1000 ES for geneset Null distribution of enrichment scores Actual ES • FDR q-value computed – corrected for gene set size and testing multiple gene sets