* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download Document
Transposable element wikipedia , lookup
Gene therapy of the human retina wikipedia , lookup
Minimal genome wikipedia , lookup
Extrachromosomal DNA wikipedia , lookup
Cre-Lox recombination wikipedia , lookup
Non-coding RNA wikipedia , lookup
Cancer epigenetics wikipedia , lookup
Human genome wikipedia , lookup
Gene nomenclature wikipedia , lookup
Deoxyribozyme wikipedia , lookup
No-SCAR (Scarless Cas9 Assisted Recombineering) Genome Editing wikipedia , lookup
DNA vaccination wikipedia , lookup
Polycomb Group Proteins and Cancer wikipedia , lookup
Genetic engineering wikipedia , lookup
Genome (book) wikipedia , lookup
Protein moonlighting wikipedia , lookup
Epitranscriptome wikipedia , lookup
Nutriepigenomics wikipedia , lookup
Epigenetics of neurodegenerative diseases wikipedia , lookup
Epigenetics of human development wikipedia , lookup
Non-coding DNA wikipedia , lookup
Genome evolution wikipedia , lookup
History of genetic engineering wikipedia , lookup
Gene expression profiling wikipedia , lookup
Vectors in gene therapy wikipedia , lookup
Designer baby wikipedia , lookup
Site-specific recombinase technology wikipedia , lookup
Primary transcript wikipedia , lookup
Genome editing wikipedia , lookup
Microevolution wikipedia , lookup
Point mutation wikipedia , lookup
Helitron (biology) wikipedia , lookup
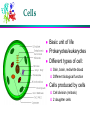
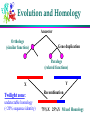
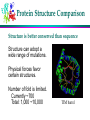
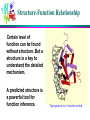
Computational Problems in Molecular Biology Dong Xu Computer Science Department 109 Engineering Building West E-mail: [email protected] 573-882-7064 http://digbio.missouri.edu Lecture Outline From DNA to gene Protein sequence and structure Gene expression Protein interaction and pathway Provide a roadmap for the entire course Biology from system level (computational perspective) About Life Life is wonderful: amazing mechanisms Life is not perfect: errors and diseases Life is a result of evolution Cells Basic unit of life Prokaryotes/eukaryotes Different types of cell: Skin, brain, red/white blood Different biological function Cells produced by cells Cell division (mitosis) 2 daughter cells DNA Double Helix (Watson & Crick) Nitrogenous Base Pairs Adenine Thymine [A,T] Cytosine Guanine [C,G] Weak bonds (can be broken) Form long chains Genome Each cell contains a full genome (DNA) The size varies: Small for viruses and prokaryotes (10 kbp-20Mbp) Medium for lower eukaryotes Yeast, unicellular eukaryote 13 Mbp Worm (Caenorhabditis elegans) 100 Mbp Fly, invertebrate (Drosophila melanogaster) 170 Mbp Larger for higher eukaryotes Mouse and man 3000 Mbp Very variable for plants (many are polyploid) Mouse ear cress (Arabidopsis thaliana) 120 Mbp Lilies 60,000 Mbp Differences in DNA ~2% ~4% ~0.2% Genes Chunks of DNA sequence that can translate into functional biomolecules (protein, RNA) 2% human DNA sequence for coding genes 32,000 human genes, 100,000 genes in tulips Gene Structure General structure of an eukaryotic gene Unlike eukaryotic genes, a prokaryotic gene typically consists of only one contiguous coding region Informational Classes in Genomic DNA Transcribed sequences (exons and introns) Messenger sequences (mRNA, exons only) Coding sequences (CDS, part of the exons only) Heads and tails: untranslated parts (UTR) Regulatory sequences ... and all the rest Identify them: gene-finding Genetic Code A=Ala=Alanine C=Cys=Cysteine D=Asp=Aspartic acid E=Glu=Glutamic acid F=Phe=Phenylalanine G=Gly=Glycine H=His=Histidine I=Ile=Isoleucine K=Lys=Lysine L=Leu=Leucine M=Met=Methionine N=Asn=Asparagine P=Pro=Proline Q=Gln=Glutamine R=Arg=Arginine S=Ser=Serine T=Thr=Threonine V=Val=Valine W=Trp=Tryptophan Y=Tyr=Tyrosine Protein Synthesis AGCCACTTAGACAAACTA (DNA) Transcribed to: AGCCACUUAGACAAACUA (mRNA) Translated to: SHLDKL (Protein) About Protein 10s – 1000s amino acids (average 300) Lysozyme sequence (129 amino acids): KVFGRCELAA AMKRHGLDNY RGYSLGNWVC AAKFESNFNT QATNRNTDGS TDYGILQINS RWWCNDGRTP GSRNLCNIPC SALLSSDITA SVNCAKKIVS DGNGMNAWVA WRNRCKGTDV QAWIRGCRL Protein backbones: Side chain Evolution of Genes: Mutation Genes alter (slightly) during reproduction Caused by errors, from radiation, from toxicity 3 possibilities: deletion, insertion, alteration Deletion: ACGTTGACTC ACGTGACTC Insertion: ACGTTGACTC AGCGTTGACTC Substitution: ACGTTGACTC ACGATGACTC Mutations are mostly deleterious Evolution and Homology Ancestor Orthologs (similar function) Gene duplication Paralogs (related functions) Y X Twilight zone: undetectable homology (<20% sequence identity) Recombination 75%X 25%Y Mixed Homology Sequence Comparison o Pairwise sequence comparison o multiple alignment SAANLEYLKNVLLQFIFLKPG--SERERLLPVINTMLQLSPEEKGKLAAV O15045 NEKNMEYLKNVFVQFLKPESVP-AERDQLVIVLQRVLHLSPKEVEILKAA P34562 KNEKIAYIKNVLLGFLEHKE----QRNQLLPVISMLLQLDSTDEKRLVMS Q06704 REINFEYLKHVVLKFMSCRES---EAFHLIKAVSVLLNFSQEEENMLKET Q92805 MLIDKEYTRNILFQFLEQRD----RRPEIVNLLSILLDLSEEQKQKLLSV O42657 EPTEFEYLRKVMFEYMMGR-----ETKTMAKVITTVLKFPDDQAQKILER O70365 DPAEAEYLRNVLYRYMTNRESLGKESVTLARVIGTVARFDESQMKNVISS Q21071 STSEIDYLRNIFTQFLHSMGSPNAASKAILKAMGSVLKVPMAEMKIIDKK Q18013 Phylogenetic Trees Understand evolution Protein Structure Lysozyme structure: ball & stick strand surface Structure Features of Folded Proteins Compact Secondary structures: loop a-helix b-sheet Protein cores mostly consist of a-helices and b-sheets Protein Structure Comparison Structure is better conserved than sequence Structure can adopt a wide range of mutations. Physical forces favor certain structures. Number of fold is limited. Currently ~700 Total: 1,000 ~10,000 TIM barrel Protein Folding Problem A protein folds into a unique 3D structure under the physiological condition Lysozyme sequence: KVFGRCELAA AMKRHGLDNY RGYSLGNWVC AAKFESNFNT QATNRNTDGS TDYGILQINS RWWCNDGRTP GSRNLCNIPC SALLSSDITA SVNCAKKIVS DGNGMNAWVA WRNRCKGTDV QAWIRGCRL Structure-Function Relationship Certain level of function can be found without structure. But a structure is a key to understand the detailed mechanism. A predicted structure is a powerful tool for function inference. Trp repressor as a function switch Structure-Based Drug Design Structure-based rational drug design is still a major method for drug discovery. HIV protease inhibitor Gene Expression Same DNA in all cells, but only a few percent common genes expressed (house-keeping genes). A few examples: (1) Specialized cell: over-represented hemoglobin in blood cells. (2) Different stages of life cycle: hemoglobins before and after birth, caterpillar and butterfly. (3) Different environments: microbial in nutrient poor or rich environment. (4) Special treatment: response to wound. Eucaryote Gene Expression Control nucleus DNA Primary RNA transcript transcriptional control Methods: Mass-spec Microarray cytosol inactive mRNA mRNA degradation control mRNA RNA processing control RNA transport control mRNA translation control protein nucleus membrane protein activity control inactive protein Gene Regulation promoter operator DNA sequence Start of transcription Microarray Experiments Regulation/function/pathway/cellular state/phenotype Disease: diagnosis/gene identification/sub-typing Microarray chip Microarray data Genetic vs. Physical Interaction Gene/protein interaction Complex system Regulatory network Physical interaction Genetic interaction Transcription factor Expressed gene Biological Pathway Studying Pathways through Systems Biology Approach RGYSLGNWVC AAKFESNFNT QATNRNTDGS TDYGILQINS RWWCNDGRTP GSRNLCNIPC sequence gene regulation structure pathway (cross-talk) function protein interaction Discussion Possible our life impacts of biotechnology to Assignments Required reading: * Chapter 13 in “Pavel Pevzner: Computational Molecular Biology - An Algorithmic Approach. MIT Press, 2000.” * Larry Hunter: molecular biology for computer scientists Optional reading: http://www.ncbi.nih.gov/About/primer/bioinformatics.html http://www.bentham.org/cpps1-1/Dong%20Xu/xu_cpps.htm