* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download Translation
Non-coding DNA wikipedia , lookup
Transfer RNA wikipedia , lookup
Genomic imprinting wikipedia , lookup
Gene nomenclature wikipedia , lookup
Transcription factor wikipedia , lookup
Long non-coding RNA wikipedia , lookup
Site-specific recombinase technology wikipedia , lookup
Genome evolution wikipedia , lookup
History of genetic engineering wikipedia , lookup
Protein moonlighting wikipedia , lookup
Polycomb Group Proteins and Cancer wikipedia , lookup
Vectors in gene therapy wikipedia , lookup
Genome (book) wikipedia , lookup
Microevolution wikipedia , lookup
Epigenetics of neurodegenerative diseases wikipedia , lookup
Epitranscriptome wikipedia , lookup
Ridge (biology) wikipedia , lookup
Non-coding RNA wikipedia , lookup
Minimal genome wikipedia , lookup
Designer baby wikipedia , lookup
Biology and consumer behaviour wikipedia , lookup
Primary transcript wikipedia , lookup
Point mutation wikipedia , lookup
Gene expression profiling wikipedia , lookup
Nutriepigenomics wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Epigenetics of human development wikipedia , lookup
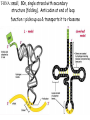
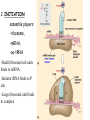
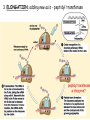
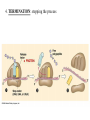
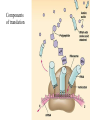
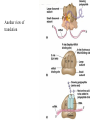
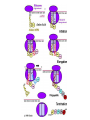
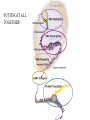
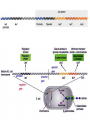
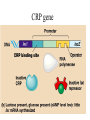
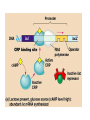
Translation • mRNA exits the nucleus through the nuclear pores • In the cytoplasm, it joins with the other key players to assemble a polypeptide. • The other parts of the machinery are: t-RNAs and ribosomes T-RNA: small, 80n, single strand with secondary structure (folding). Anticodon at end of loop. function = picks up aa & transports it to ribosome Ribosomes: composed of rRNA and proteins Sequence of 4 Steps in Translation... 1. ACTIVATION : Add an amino acid to tRNA Requires enzyme and ATP Creates an aa-tRNA 2. INITIATION: assemble players -ribosome, -mRNA, -aa-tRNA -Small ribosomal sub units binds to mRNA. -Initiator tRNA binds to P site -Large ribosomal unit binds to complex 3. ELONGATION: adding new aa’s - peptidyl transferase 4. TERMINATION: stopping the process Components of translation Another view of translation PUTTING IT ALL TOGETHER! REGULATION OF GENE EXPRESSION •The control in the DNA transcription process is very tight. •Cells are able to "turn on" or "turn off" genes when their products are not required in cell metabolism or control. • Regulation of gene expression is now only being to be fully understood and is a major area of research today. GENE REGULATION IN PROKARYOTES • Genes for a particular metabolic pathway are usually in clusters, under the control of one promoter and one operator. • This cluster of genes, with its regulatory sites is called an operon The lac operon • E. coli can use lactose as a source of energy • To use lactose, it must split the lactose into glucose and galactose. This means producing enzymes. • Only produces enzymes when lactose is present in the environment. WHY? • Three genes are involved: 1. LacZ: codes for B-galactosidase: degrades lactose 2. LacY: codes for B-galactosidase permease: causes lactose to enter the cell 3. LacA: codes for a transacetylase: function unknown! • The lacl gene codes for the lacl protein • This protein is always produced. • When lactose is NOT present in the environment, the lacl protein binds to the operator site. • This blocks RNA polymerase from attaching to the promoter site. Transcription is turned OFF operator - binds repressor protein • When lactose is PRESENT in the environment, The lactose binds to the lacl protein, and changes its shape • The lacl protein “falls” off the operator site and RNA polymerase can now attach to the promoter site and transcription of the lac genes proceeds. • Lactose is an inducer molecule. Its presence activates transcription of the genes that degrade it. • Often, both glucose and lactose are present in the environment. • E. coli uses glucose first, since the enzymes for its use are always present. • There is a mechanism to slow down the use of lactose even if it is present. • When all glucose is used, then transcription of lac genes will speed up. •CATABOLIC REPRESSION •glucose prevents the action of the LAC operon through another regulator-like protein, the cAMP receptor protein (CRP) • CRP binds to DNA at the CRP gene •CRP is aka as CAP ( Catabolite Activator protein) •This involves the use of cAMP as an intermediate messenger CRP gene • CRP (aka CAP) is an allosteric protein, regulated by cAMP • when glucose is high - lots of ATP & little cAMP • CRP-alone conformation doesn't bind to CRP DNA region - favors slow transcription of lac genes • when glucose is low - all the ATP is hydrolyzed favoring high cAMP amounts • cAMP-CRP conformation can bind to CRP DNA region -favors rapid transcription of lac genes The TRP operon • This cluster of genes is responsible for the enzymes that synthesize Tryptophan, an essential amino acid. • They are always turned on EXCEPT when tryptophan is present in the environment. • The operon consist of five genes, an operator, a promoter and the trp repressor protein. • Tryptophan is needed to inactivate the trp operon—it is a corepressor. • This type of regulation is by repression because the effector molecule interacts with the repressor protein so that it can bind to the operator