* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download מצגת של PowerPoint - Tel Aviv University
Minimal genome wikipedia , lookup
Nutriepigenomics wikipedia , lookup
History of genetic engineering wikipedia , lookup
Genomic imprinting wikipedia , lookup
Gene desert wikipedia , lookup
Neuronal ceroid lipofuscinosis wikipedia , lookup
Gene therapy wikipedia , lookup
Biology and consumer behaviour wikipedia , lookup
Vectors in gene therapy wikipedia , lookup
Epigenetics of human development wikipedia , lookup
Point mutation wikipedia , lookup
Gene nomenclature wikipedia , lookup
Copy-number variation wikipedia , lookup
Therapeutic gene modulation wikipedia , lookup
Polycomb Group Proteins and Cancer wikipedia , lookup
X-inactivation wikipedia , lookup
Gene expression profiling wikipedia , lookup
Genome editing wikipedia , lookup
Genome (book) wikipedia , lookup
Gene expression programming wikipedia , lookup
Protein moonlighting wikipedia , lookup
Designer baby wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Site-specific recombinase technology wikipedia , lookup
Genome evolution wikipedia , lookup
Nicotinamide adenine dinucleotide wikipedia , lookup
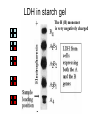
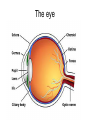
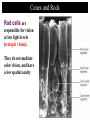
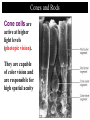
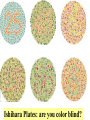
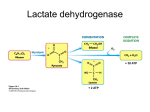
4: Genome evolution Types of Genomic Duplications •Part of an exon or the entire exon is duplicated •Complete gene duplication •Partial chromosome duplication •Complete chromosome duplication •Polyploidy: full genome duplication After a Gene is Duplicated Alternative fates: 1. It can die and become a pseudogene. 2. It can retain its original function, thus allowing the organism to produce double the amount of the derived protein. 3. The two copies can diverge and each one will specialize in a different function. Divergence One copy dies Identical copies Isozyme (LDH) Complete gene duplication and different function Human L-lactate dehydrogenase M4 (the isozyme found in skeletal muscle). Isozyme Isozymes are enzymes that catalyze the same biochemical reaction but may differ from one another in tissue specificity, developmental regulation or biochemical properties. Isozymes are encoded by different loci, usually duplicated genes, as opposed to allozymes, which are different alleles of an enzyme. Isozymes are important for development. They allow differentiation of cells. Lactate dehydrogenase (LDH) An enzyme that is composed of two types of subunits, M and H (formally A and B), each encoded by a different gene. “M” stands for “muscle” and “H” for heart. •The M subunit is encoded by LDHA, located on chromosome 11p15.4 •The H subunit is encoded by LDHB, located on chromosome 12p12.2p12.1 (partial) LDH alignment Lactate dehydrogenase (LDH) The protein itself is tetrametric. It can have one of the following isozymes: •H4 (also called LDH1) •H3M (also called LDH2) •H2M2 (also called LDH3) •HM3 (also called LDH4) •M4 (also called LDH5) Expressed mostly in the heart + blood cells Expressed mostly in white blood cells Expressed mostly in the lung Expressed mostly in the kidney, placenta Expressed mostly in the liver + muscles LDH in starch gel The H (B) monomer is very negatively charged Lactate dehydrogenase (LDH) The enzyme catalyzes the following biochemical reaction: Lactate dehydrogenase (LDH) The enzyme catalyzes the following biochemical reaction: Pyruvate + NADH Lactate + NAD How to color: we give to the enzyme, a buffer sullution with lactic acid, NAD, NBT (Nitro blue tetrazolium) and PMS (phenazine methosulfate). LDH Lactate + NAD NADH + H+ + PMS NBT + PMS[reduced] Pyruvate + NADH NAD+ + PMS[reduced] NBT[reduced] + PMS The NBT is dark blue when it is reduced Lactate dehydrogenase (LDH) • The M form (LDH-A) is found predominantly in white skeletal muscle (fast twitch glycolytic fibers) and is best suited for pyruvate reduction in anaerobic conditions. • the H form (LDH-B) is found in more aerobic tissues such as heart and brain and is most efficient for lactate oxidation. Lactate dehydrogenase (LDH) The reaction in context: Glycolyses Glucose + 2 ADP + 2 NAD+ + 2 Pi -----> 2 Pyruvate + 2 ATP + 2 NADH + 2 H+ Oxidative Phosphorylation (mitochondria) NADH+ 3 ADP + 3 Pi -----> NAD+ ++H+ + 3ATP Later Pyruvate + ADP + NAD-----> CO2 + NADH + ATP When we run for our lives, there’s not enough NAD here, So LDH (M4 TYPE) in muscles does the following reaction (when there’s lack of oxygen) Pyruvate + NADH Lactate + NAD Lactate dehydrogenase (LDH) The lactate diffuses to the blood, and from their to organs where there’s no oxygen missing (the heart). There, not much of glucose is needed, there’s enough NAD, and the opposite reaction is catalyzed, by LDH type H4 Pyruvate + NADH Lactate + NAD Lactate dehydrogenase (LDH) When a cell dies its LDH is released to the blood. LDH is one of the main components in blood tests. High level of LDH are indicative of heart attacks, cancer or anemia. If LDH levels are high, another test is performed to test the level of the different isozymes, so that the source of the problem can be more accurately inferred. Lactate dehydrogenase (LDH) This is an example in which two duplicated genes have become specialized to different tissues. The isozymes are also differentially expressed in different developmental stages. Before birth the heart is more anaerobic compared with adulthood. Indeed, before birth the main isozyme in the heart is the M4, and with time it switches to HM3 (at birth), to H2M2 and HM3 at 1 year after birth, and to H3M AND H4 after 2 years. My main LDH is HM3. Great! My main LDH is HM3 LDH of mices B4 AB3 A2B2 A3B A4 -9 -5 -1 0 +12 +21 -9, -5, -1 days before birth as well as +12 and +21 days after birth and in adult mice Lactate dehydrogenase (LDH) 4: Genome evolution Vision. The Opsins stories Complete gene duplication and different function The eye Retina Cones and Rods There are two types of photoreceptor cells in the human retina, rods and cones. Cones and Rods Rod cells are responsible for vision at low light levels (scotopic vision). They do not mediate color vision, and have a low spatial acuity Cones and Rods Cone cells are active at higher light levels (photopic vision). They are capable of color vision and are responsible for high spatial acuity Colors There are 3 types of pigments in cones. Their peaks of absorption are at about 430, 530, and 560 nanometers. Colors The cones are “loosely” called "blue", "green", and "red“. “loosely” because: 1. the names refer to peak sensitivities (which in turn are related to the ability to absorb light) rather than to the way the pigments would appear if we were to look at them. Colors The cones are “loosely” called "blue", "green", and "red“. “loosely” because: 2. Monochromatic lights whose wavelengths are 430, 530, and 560 nanometers are not blue, green, and red but violet, blue-green, and yellow-green Terminology Terminology is almost impossible to change. Some call the cones just long, middle, and short. An impossible elephant Opsin and retinal Each photopigment consists of two parts: a protein called opsin and a lipid derivative called retinal. The opsin is a member of the superfamily of Gprotein coupled receptors. The opsin’s sequence determines the absorbance Opsin Genetics The blue opsin is encoded by an autosomal gene. The red and green opsins are encoded by X-linked genes. There are cases in which the green opsin is duplicated on the X chromosome. Red and green are very similar in amino-acid sequence (96%). Blue is more diverged (43%). Blue diverged about 500 mya (million years ago). Red and green diverged only 25-35 mya. Opsin Genetics Indeed new-world monkeys have only one X-linked pigment, so the divergence of green and red occurred after the divergence of new-world monkeys from old-world monkeys. Thus, old-world monkeys are trichromatic. Opsin Genetics New-world monkeys have only one X-linked locus (except for the howler monkey from the Alouatta genus). howler monkey Opsin Genetics But the X-linked locus in the new world monkeys, such as for squirrel monkeys and tamarins, can be highly polymorphic, with some alleles similar to the red opsion and some to the green. Thus, a female can be trichromatic (if heterozygous) but males are always dichromatic. Dichromatic monkeys cannot distinguish between red and green. Squirrel monkeys Ishihara Plates: are you color blind? The individual with normal color vision will see the number 5 revealed in the dot pattern. An individual with Red/Green (the most common) color blindness will see the number 2 revealed in the dots 5 or 2? Opsin: conclusion one Old world monkeys achieved trichomatic vision by a mechanism akin to isozymes (different proteins coded by different loci). Heterozygous female squirrel monkeys achieved trichomatic vision by using two “allozymes” (distinct proteins encoded by different allelic forms at a single locus). The polymorphism is probably a form of overdominant selection. Opsin: selective advantage? The selective advantage of trichromatic vision is thought to be the ability to detect ripe fruits against a background of dense green foliage. Pheromones Old world monkeys, apes and humans have trichromatic vision. Other placental mammals don’t. Hypothesis: The development of trichromatic vision led to the eventual deterioration of pheromones sensitivity. This is explained by the superiority of trichromatic vision over pheromones as a means of communication. Against this hypothesis: Howler monkeys communicate via both channels. 4: Genome evolution Carbamoyl-phosphate synthetase Carbamoyl-phosphate synthetase 2MgATP + HCO3- + Glutamine + H2O CPS 2MgADP + Pi + Glutamate + Carbamoyl-P Carbamoyl-phosphate synthetase The reaction is needed in a few places: 1. Pyrimidine biosynthesis. 2. Arginine biosynthesis. 3. Urea cycle (amino acid catabolism). Carbamoyl-phosphate synthetase In several organisms the CPS enzyme was duplicated. In these organisms, the enzymes are always specialized to one of these reactions. When there’s no duplication – the enzyme does all 3 functions (multifunctional). From the specific example to the general theory Two theories of gene duplication: 1. After the gene was duplicated, there was a period of random mutations, until the gene obtained a new function, and from this point onwards, selection is operating on it. The orthodox model of gene duplication The orthodox model for the evolution of functionally novel proteins (Ohno 1970 & Kimura 1983) After gene duplication, one copy is rendered redundant and can accumulate substitutions at random. By chance, some of these substitutions may result in a new function. Difficulties with the orthodox theory One difficulty with this theory is that: Unless the new function can be acquired through one or a few nucleotide substitutions, it is more than likely that the copy will become a pseudogene rather than a new functional gene. Difficulties with the orthodox theory Another problem with this theory is that: There are many evidences of positive selection after gene duplication (rather than relaxation of functional constraints). From the specific example to the general theory The second theory is: 2. Genes that survive after duplication have had more than one function before. After selection, there’s a specialization of the two genes. This gives a selective advantage for the duplication. From the specific example to the general theory The CPS example, clearly supports the second theory.