* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download Dot Plot - APBioNET
Vectors in gene therapy wikipedia , lookup
Zinc finger nuclease wikipedia , lookup
Biochemistry wikipedia , lookup
Gene expression wikipedia , lookup
Nucleic acid analogue wikipedia , lookup
Transposable element wikipedia , lookup
Endogenous retrovirus wikipedia , lookup
Deoxyribozyme wikipedia , lookup
Molecular ecology wikipedia , lookup
Biosynthesis wikipedia , lookup
Bisulfite sequencing wikipedia , lookup
Promoter (genetics) wikipedia , lookup
Silencer (genetics) wikipedia , lookup
Multilocus sequence typing wikipedia , lookup
Non-coding DNA wikipedia , lookup
Genetic code wikipedia , lookup
Two-hybrid screening wikipedia , lookup
Community fingerprinting wikipedia , lookup
Protein structure prediction wikipedia , lookup
Point mutation wikipedia , lookup
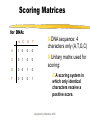
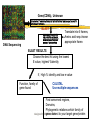
Part I : SEQUENCE COMPARISON PAIRWISE ALIGNMENT Manisha Brahmachary designed by Manisha, NUS OUTLINE What is sequence Comparison Ways to do Sequence Comparison Dot Plot BLAST FASTA designed by Manisha, NUS What is sequence alignment or sequence comparison? Given two sequences of letters and a scoring scheme for evaluating matching letters , find best pairing from one sequence to letters of the other sequence. THIS IS A RATHER LONGER SENTENCE THAN THE NEXT. THIS IS A SHORT SENTENCE Align: THIS IS A RATHER LONGER SENTENCE THAN THE NEXT. THIS IS A#######SHORT###SENTENCE############## (path 1) or THIS IS A SHORT#########SENTENCE############## (path 2) designed by Manisha, NUS Aligning biological sequences DNA (4 letter alphabet) TTGACAC TTTACAC Proteins (20 letter alphabet) RKVA--GMAKPNM RKIAVAAASKPAV designed by Manisha, NUS Why do Sequence Alignment? Finding novel genes in silico Phylogenetic/Evolutionary Structure-template for modelling Functional prediction designed by Manisha, NUS Types of Sequence Comparison Pairwise Alignment Comparison of two sequences Multiple Alignment Comparison of more than two sequences designed by Manisha, NUS CONCEPTS IN SEQUENCE COMPARISON IDENTITY Percentage identity between sequences means that they have a certain number of residues (nucleotide /amino- acids ) that are identical at that particular position after aligning both sequences. designed by Manisha, NUS Query: RCI CTRGFCRCLCRR Subject: RCLCRRGVCRCICT R • Exact match (shown by | ) : 10 identical residues • Above example : •Percentage identity: 10 identical matches /15 residues in the aligned sequence *100 = 66% identity designed by Manisha, NUS Query: RCI CTRGFCRCLCRR Subject: RCLCRRGVCRCICT R MISMATCH(s) HERE designed by Manisha, NUS Query: RCICT-RGFCRCLC---RR Subject: RCLCRRGVCRCICTAR • Mismatch when different characters , therefore insertion of gaps. • Gaps have penalties: • Insertion of first gap( GAP OPENING) : high penalty (For eg. –2, subtracting 2 ) • Insertion of consecutive gaps ( GAP EXTENSION): less penalty (For eg. -1 (subtracting 1 for each consecutive gap) •More no. of gaps lesser the score of the alignment designed by Manisha, NUS RCICT-RGFCRCLC---RR RCLCRRGVCRCICTAR- Substitution: Less score than identical match For eg: +1 per substitution designed by Manisha, NUS Substitution - Replace a residue with another of similar physiochemical property. Category Amino Acid Acids and Amides Asp (D) Glu(E) Asn (N) Gln (Q) Basic His (H) Lys (K) Arg (R) Aromatic Phe (F) Tyr (Y) Trp (W) Hydrophilic Ala (A) Cys (C) Gly (G) Pro (P) Ser (S) Thr (T) Hydrophobic Ile (I) Leu (L) Met (M) Val (V) designed by Manisha, NUS Similarity RCICT-RGFCRCLC---RR RCLCRRGVCRCICTAR Similarity = Identical matches + Substitutions Eg. (10 identical matches + 2 substitution) / 15 aligned residues * 100 = 80% similarity designed by Manisha, NUS For DNA: Identity and gap are applicable ACTCGGCCCCGCG CTCACTG C ACTCGGAC - - GCG CTCAGTGC designed by Manisha, NUS Similarity Vs. Homology Homology: When two similar proteins come from a common ancestor. Homology is inferred from Similarity If two sequences are similar, then they are known as homologous sequences. Usually, at least 30% identity over 400 bp for DNA sequences and over 125 amino acids for proteins. designed by Manisha, NUS Scoring Matrices used in sequence comparison What is a scoring Matrix: Scoring matrices are used when we compare sequences with one another Gives us a measure of which residue can be substituted by which residue. designed by Manisha, NUS Scoring Matrices For Amino acids, Each amino acid is compared to every other and a score is given to this pair High score if they are the same residue (e.g. Cysteine compared to cysteine) Low, if they are very different (e.g. Tryptophan compared to cysteine) designed by Manisha, NUS Scoring Matrices for DNA: A C G T A 1 0 0 0 C 0 1 0 0 G 0 0 1 0 T 0 0 0 1 DNA sequence: 4 characters only (A,T,G,C) Unitary matrix used for scoring: A scoring system in which only identical characters receive a positive score. designed by Manisha, NUS SCORING SCHEMES FOR PROTEIN SEQUENCE ALIGNMENTS Scoring matrices used are: PAM(Point Accepted Mutation) and BLOSUM(BLOcks SUbstitution Matrix BLOSUM45---->BLOSUM 90 means MORE DIVERSE TO LESS DIVERSE PAM30---PAM250 means LESS DIVERSE TO MORE DIVERSE NOTE: Many different matrices are in use, each gives different values to pairs of amino acids Depending on how distantly related your sequences are, you might want to choose different matrices for your comparisons designed by Manisha, NUS Scoring Matrices Notes: BLOSUM 45 PAM250 BLOSUM62 BLOSUM90 PAM160 PAM 100 MORE DIVERGENT LESS DIVERGENT designed by Manisha, NUS Ways to do Pairwise Alignment Dot Plot (simplest method) Statistical computation based Local alignment e.g. BLAST, FASTA Global alignment e.g. CLUSTAL designed by Manisha, NUS What are Dot Plots Program to do sequence comparison to find out: –Are the two sequences similar ? – Are there Repeat regions in your sequence? designed by Manisha, NUS STEPS IN DOT PLOT •Take two sequences to be compared •Sequence A:MEHRKPGTGQ •Sequence B:MEHRKPGTGQ •Place sequence A in x-axis (Row). Place sequence B in yaxis (Column) M E H R K P G T G Q X-axis Y-axis designed by Manisha, NUS •Plot a dot everytime there is a match between an element of row sequence and an element of column sequence •Do you see any diagonal line extending? •If yes, then there is a match ! designed by Manisha, NUS Patterns in Dot Plot GGTCCTTGGCTGAAAGACCCCA When two sequences are “identical” Sequence : GGTCCTTGGCTGAAAG ACCCCA designed by Manisha, NUS Application of Dot Plot Using self comparison : Finding Repeats Sequence used: CATCTCAAAAACAACAACAAAAAAAAAAAAAAAAGAAAAAAAA Human ALU sequence CATCTCAAAAACAACAA CAAAAAAAAAAAAAAAA GAAAAAAAA •Omit main diagonal •Clusters of diagonal lines show repeats in the sequence. designed by Manisha, NUS Notes:What are repeats? Repeats:are stretches of repeated regions of residues in a sequence. Importance of repeats: In protein: Regulatory regions Binding sites In DNA: Present in Transposons, chromosomal mutational hotspots, many genetic diseases related with repeats.eg.Huntington. designed by Manisha, NUS Patterns in Dot Plot When two sequences are similar : Broken diagonal,the interrupted region shows regions of mismatch designed by Manisha, NUS Patterns in Dot Plot Two different, but related sequences GREGYPADSKGCKITCFLTAAGYCNTECTLKKGSSGYCAWPA Broken diagonal clusters of dots parallel to the central diagonal. Distance between the lines show no. of insertions done to get the alignment. designed by Manisha, NUS Two models of alignment: Local and Global alignments Global alignment: Looks for similarity across full extent of sequences Site:http://www2.igh.cnrs.fr/bin/align-guess.cgi designed by Manisha, NUS GLOBAL Alignment The two sequences are matched across their whole sequence length. designed by Manisha, NUS Local alignment Looks for regions of similarity in parts of the sequences only Softwares : BLAST, FASTA designed by Manisha, NUS Local Alignment Example of local alignment between two sequences using lalign program. (http://www.ch.embnet.org/software/LALIG N_form.html) Notice that the alignment is shown only of those regions that have strong identity or strong similarity designed by Manisha, NUS Why two different models? Global alignment High degree of Homology Good for modelling Local Alignment Localised Similarity ( conserved regions with structural , functional importance, Repeats, Domains) designed by Manisha, NUS FASTA Fast Alignment (expanded form of FASTA)by Pearson and Lipmann. Is a method based on dynamic programming. Websites available: http://www.ebi.ac.uk/fasta33/ http://www.dna.affrc.go.jp/htdocs/Blast/fasta designed by Manisha, NUS .html What is BLAST? Basic Local Alignment Search Tool (BLAST) Method for Pairwise Alignment. Is used to search for homologous sequences from a database (of nucleotide/protein sequence) for a given query sequence. Modified version of FASTA Faster in generating output. Sites for doing BLAST: http://www.ncbi.nlm.nih.gov designed by Manisha, NUS How to go about doing BLAST SARS virus gene: SGFRKMAFPSGKVEGCMVQVTCGTTTLNGLWLDDTVYCPRHVICTAEDMLNPNYE DLLIRKSNHSFLVQAGNVQLRVIGHSMQNCLLRLKVDTSNPKTPKYKFVRIQPGQTF SVLACYNGSPSGVYQCAMRPNHTIKGSFLNGSCGSVGFNIDYDCVSFCYMHHMEL PTGVHAGTDLEGKFYGPFVDRQTAQAAGTDTTITLNVLAWLYAAVINGDRWFLNRF TTTLNDFNLVAMKYNYEPLTQDHVDILGPLSAQTGIAVLDMCAALKELLQNGMNGR T ILGSTILEDEFTPFDVVRQCSGVTFQ designed by Manisha, NUS designed by Manisha, NUS BLAST output for a protein query sequence from a SARS virus Score (bits) is the score given letter by letter during alignment based on the Subtitution matrices. High score = less E value. designed by Manisha, NUS •E value: No. of chance alignments that one will get as hits. •Lower the E value lesser no. of chance hits •E value of zero or less than zero indicates very good hit (highly homologous sequence) •E value is also known as P(N) in some BLAST programs designed by Manisha, NUS BLAST OUTPUT Gives the identity Gives the similarity designed by Manisha, NUS BLAST BLAST query schemes: Amino acid seq: against db? Blastp (protein sequence db) Tblastn (translated nucleotide sequence db) DNA seq: against db? Blastn (nucleotide db) Blastx ( protein sequence db) Tblastx (translated nucleotide sequence db) designed by Manisha, NUS Gene(CDNA), Unknown CTAACATGCTTAGGATAATGGCCTCTCTTGTTCTTGCTCGCAAACATAACACTT GCTGTAACTTATCACA BLAST NMLRIMASLVLARKHNTC CNLSHRFYRLANECAQVL SEMVMCGGSLYVKPGGT SSGDATTAYANSVFNIC DNA Sequencing Translate into 6 frames, Amino acid seq.choose appropriate frame. BLAST RESULTS Choose the best hit using the lowest E value, highest %identity If , High % identity and low e-value Function, family of gene found CLUSTAL Use multiple sequences Find conserved regions, Domains, Phylogenetic relations:which family of designed by Manisha, NUS gene closest to your target gene/protein SUMMARY TODAY WE LOOKED AT: Methods to compare two sequences: Dot plots (simplest, graphical view) Different patterns of Dot plots Local alignment Global alignment Difference between these two models FASTA BLAST other types of BLAST designed by Manisha, NUS