* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download Slides - WordPress.com
Fatty acid synthesis wikipedia , lookup
Gaseous signaling molecules wikipedia , lookup
Biosynthesis wikipedia , lookup
Mitogen-activated protein kinase wikipedia , lookup
Electron transport chain wikipedia , lookup
Biochemistry wikipedia , lookup
Cyanobacteria wikipedia , lookup
Nicotinamide adenine dinucleotide wikipedia , lookup
Metalloprotein wikipedia , lookup
Paracrine signalling wikipedia , lookup
Adenosine triphosphate wikipedia , lookup
Light-dependent reactions wikipedia , lookup
Amino acid synthesis wikipedia , lookup
Sulfur cycle wikipedia , lookup
Biochemical cascade wikipedia , lookup
Photosynthesis wikipedia , lookup
Oxidative phosphorylation wikipedia , lookup
Citric acid cycle wikipedia , lookup
Evolution of metal ions in biological systems wikipedia , lookup
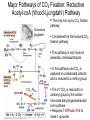
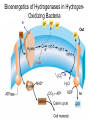
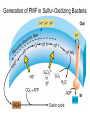
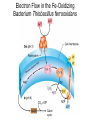
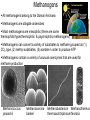
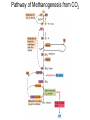
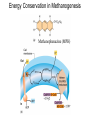
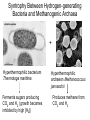
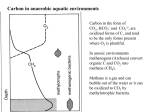
Lecture #18: Something from Almost Nothing: Chemoautotrophy and Chemolithotrophy Background Reading for Lecture #18 Physiology & Biochemistry of Prokaryotes 3rd Ed.: Ch. 12, pp. 345-356; Ch. 13, pp. 367-374 Brock Biology of Microorganisms 11th Ed: Ch.17, pp. 548-555, 583-585, 560-562, 564-568, 575-577 Brock Biology of Microorganisms 12th ed.: Ch. 20, pp. 591-602, 622-623, 627-635 Article(s) to be discussed Berg review (Autotrophic carbon fixation) Topics: Chemoautotrophy Hydrogen Bacteria Sulfur-oxidizing Bacteria Sulfur-reducing Bacteria Iron-oxidizing Bacteria Methanogenesis Methanogenesis and Syntrophy Chemolithotrophy Supports Chemoautotrophy & NAD(P)H Chemolithotrophy ability of microbes to obtain energy from the oxidation of inorganic compounds Chemoautotrophy ability of microbes to use CO2 as a sole source of carbon Major Pathways of CO2 Fixation: CalvinBenson-Bassham (CBB) Pathway CBB is most common method of CO2 fixation on Earth CBB is only used by Bacteria and Eukaryota The CBB pathway requires 3 ATP for the fixation of 1 CO2 (net result of CBB is formation of 1 triose-P from 3 CO2 at the expense of 9 ATP & 6 NAD(P)H) RuBisCO (ribulose-1,5bisphosphate carboxylase/oxygenase is the key enzyme of the pathway and catalyzes the carboxylation or oxygenation of RuBP Major Pathways of CO2 Fixation: Reductive Tricarboxylic Acid (rTCA) Cycle Chlorobium chlorochromatii Frd Light driven rxns provides reduced Fd Acl Pyruvate synthase Oor rTCA cycle was originally discovered in green sulfur phototrophs and has since been identified in a variety of chemoautotrophs rTCA cycle specific enzymes are 2-oxoglutarate:ferredoxin oxidoreductase (Oor), fumarate reductase (Frd), and ATP citrate lyase (Acl) rTCA cycle pathway tends to be in organisms that live in low O2 Major Pathways of CO2 Fixation: 3hydroxypropionate (3-HP) Cycle Originally identified from the anaerobic phototroph Chloroflexus Chloroflexus Key pathway enzymes are acetyl-coA/propionyl-coA carboxylase, malonyl-coA reductase, propionyl-coA synthase An additional enzyme 4hydroxybutyryl-coA dehydratase was identified in some Archaea Requires 7 ATP and 6 NAD(P)H Most microbes that use the 3-HP cycle can also grow heterotrophically Major Pathways of CO2 Fixation: Reductive Acetyl-coA (Wood-Ljungdahl) Pathway The only non-cyclic CO2 fixation pathway Clostridium ljungdahlii Considered the first evolved CO2 fixation pathway This pathway is only found in anaerobic chemoautotrophs In this pathway one CO2 is captured on a dedicated cofactor and is reduced to a methyl group pyruvate CO2, Fdred The 2nd CO2 is reduced to a carbonyl group by the carbon monoxide dehydrogenase/acetylcoA synthase Requires 1 ATP and 3 Fd to make 1 pyruvate * * * * Chemolithotrophic Options to Support Chemoautotrophy Hydrogen oxidation Sulfur oxidation Metal oxidation (Fe, Mn, As, Cu) Ammonia oxidation Hydrogen Bacteria: Oxidation of H2 to Generate Energy Capable of growing with H2 as the only e- donor and O2 as the eacceptor H2 + ½ O2 H2O G0 = -237 kJ Many hydrogen bacteria can grow autotrophically Ralstonia eutropha, Gram negative bacteria All hydrogen bacteria contain one or more hydrogenases to generate ATP or produce reducing power Most hydrogen bacteria are considered to be facultative chemolithotrophs Aquifex Sp. marine hyperthermophile Bioenergetics of Hydrogenases in HydrogenOxidizing Bacteria H+ NADH + 2H+ Sulfur-Oxidizing Bacteria H2S + 2 O2 SO42- + 2 H+ G0 = -798.2 kJ HS- + ½ O2 + H+ S0 + H2O -209.4 kJ S0 + H2O 1 ½ O2 SO42- + 2 H+ -587.1 kJ S2O32- + H2O + 2 O2 2 SO42- + 2 H+ -818.3 kJ Deposition of internal sulfur granules by Beggiatoa Sulfur spring colonized with Sulfolobus Sp. Attachment of the sulfur-oxidizing archaeon Sulfolobus acidocaldarius to a crystal of elemental sulfur Oxidation of Reduced Sulfur Species by Sulfur Chemolithotrophs Generation of PMF in Sulfur-Oxidizing Bacteria Iron-Oxidizing Bacteria + 0.77 V Iron oxidation (Fe2+ Fe3+) releases only a small amount of energy ½ O2 H2O (+ 0.82) At neutral pH under oxic conditions, Fe will be abiologically oxidized Fe(OH)3 Most Fe-oxidizing bacteria live in acidic environments Empty iron-encrusted sheaths of Sphaerotilus collected from seepage at the edge of a small swamp Some Fe-oxidizing bacteria such as Thiobacillus ferrooxidans are autotrophic Thiobacillus ferrooxidans Electron Flow in the Fe-Oxidizing Bacterium Thiobacillus ferrooxidans Methanogens All methanogens belong to the Domain Archaea Methanogens are obligate anaerobes Most methanogens are mesophilic (there are some thermophilic/hyperthermophilic & psychrophilic methanogens) Methanogens can convert a variety of substrates to methane grouped as (1) CO2 type, (2) methyl substrates, (3) acetate in order to produce ATP Methanogens contain a variety of unusual coenzymes that are used for methane production Methanococcus janaschii Methanosarcina barkeri Methanobacterium Methanothermus thermoautotrophicum fervidus Coenzymes of Methanogenic Archaea Coenzymes of Methanogenic Archaea Continued Pathway of Methanogenesis from CO2 Energy Conservation in Methanogenesis Methanophenazine (MPH) Methanogenesis and Syntrophy Ethanol Acetate Methane +19 kJ/rxn -137 kJ/rxn -111.3 kJ/rxn Syntrophy is also referred to as interspecies H2 transfer Syntrophy Between Hydrogen-generating Bacteria and Methanogenic Archaea + Hyperthermophilic bacterium Thermotoga maritima Hyperthermophilic archaeon Methanococcus jannaschii Ferments sugars producing CO2 and H2, (growth becomes inhibited by high [H2]) Produces methane from CO2 and H2