* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download Chapter 22 Biosynthesis of amino acids, nucleotides and related
Enzyme inhibitor wikipedia , lookup
Microbial metabolism wikipedia , lookup
Nucleic acid analogue wikipedia , lookup
Oligonucleotide synthesis wikipedia , lookup
Adenosine triphosphate wikipedia , lookup
Nitrogen cycle wikipedia , lookup
Point mutation wikipedia , lookup
Photosynthetic reaction centre wikipedia , lookup
Ribosomally synthesized and post-translationally modified peptides wikipedia , lookup
Catalytic triad wikipedia , lookup
Evolution of metal ions in biological systems wikipedia , lookup
Fatty acid metabolism wikipedia , lookup
Fatty acid synthesis wikipedia , lookup
Protein structure prediction wikipedia , lookup
Oxidative phosphorylation wikipedia , lookup
Proteolysis wikipedia , lookup
Metalloprotein wikipedia , lookup
Peptide synthesis wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Genetic code wikipedia , lookup
Citric acid cycle wikipedia , lookup
Biosynthesis of doxorubicin wikipedia , lookup
Biochemistry wikipedia , lookup
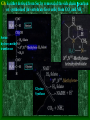
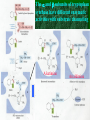
Chapter 22 Biosynthesis of amino acids, nucleotides and related molecules 1. Reduction (fixation) of N2 into ammonia (NH3 or NH4+) 2. Synthesis of the 20 amino acids. 3. Synthesis of other biomolecules from amino acids 4. The de novo pathways for purine and pyrimidine biosynthesis. 5. The salvage pathways for purine and pyrimidine reuse. 1. The nitrogenase complex in certain bacteria (diazotrophs,固氮生物) catalyzes the conversion of N2 to NH3 • The nitrogen in amino acids, purines, pyrimidines and other • • • • biomolecules ultimately comes from atmospheric nitrogen. Cyanobacteria (蓝藻细菌, photosynthetic) and rhizobia (根 瘤菌, symbiont) can fix N2 into NH3. The reduction of N2 to NH3 is thermodynamically favorable : N2 + 6e- + 6H+ 2NH3 G`o = -33.5kJ/mol But kinetically unfavorable: the bond energy for the triple bond in N2 is 942 kJ/mol. • The nitrogenase (固氮酶) complex mainly consists of two types of enzymes: the dinitrogenase and the dinitrogenase reductase. • The dinitrogenase (containing molybdenum,钼, thus called the MoFe protein) is a tetramer of two different subunits, containing multiple 4Fe-4S centers and two Mo-Fe clusters. • The dinitrogenase reductase (also called the Fe protein) is a dimer of two identifcal subunits, containing a single 4Fe-4S redox center. • The nitrogenase complex is highly conserved among different diazotrophs. Cyanobacteria and Rhizobia can fix N2 into ammonia Rhizobia exist in nodules of leguminous plants The nitrogenase complex The dinitrogenase reductase (dimer) The dinitrogenase (tetramer) The dinitrogenase reductase (dimer) 3Fe-3S Fe-Mo cofactor 3Fe-3S Mo ADP 4Fe-4S 4Fe-4S ADP 4Fe-4S (P-cluster) 2. Electrons are transferred through a series of carriers to N2 for its reduction on the nitrogenase complex • Eight electrons are believed to be needed for each round of fixation reaction: with six for reducing one N2 and two for reducing 2 H+ (to form H2). • The electrons mainly come from reduced ferredoxin (from photophosphorylation) or reduced flavodoxin (from oxidative phosphorylation) and are transferred to dinitrogenase via dinitrogenase reductase. • For each electron to be transferred from dinitrogenase reductase to dinitrogenase, two ATPs are hydrolyzed causing a conformational change which reduces the electron affinity for the reductase (i.e., an increased reducing power). • The oxidized and reduced dinitrogenase reductase dissociates from and associates with the dinitrogenase, respectively. • The nitrogenase is in imperfect enzyme: H2 is formed along with NH3. • The overall reaction catalyzed is: • N2 + 8H+ +8e- + 16ATP + 16H2O • 2NH3 + H2 + 16ADP + 16Pi Electrons are transferred to N2 bound in the active site of dinitrogenase via ferredoxin/flavodoxin and dinitrogenase reductase 3Fe-3S 3Fe-3S N2 is believed to bind to the cavity of the Fe-Mo cofactor of the dinitrogenase active site. 3. The nitrogenase complex is extremely labile to O2 and various protective mechanisms have evolved • Some diazotrophs exist only anaerobically. • Some cyanobacterial cells develop thick walls to prevent O2 from entering. • The bacteria in root nodules are isolated from O2 by being bathed in a solution of the oxygen-binding protein leghemoglobin. Leghemoglobin, produced in legume plants, has a high affinity to O2 and protects the nitrogenase complex in rhizobia 4. Reduced nitrogen in the form of + NH4 is assimilated into amino acids via a two-enzyme pathway • First NH4+ is added to the side chain of glutamate to form glutamine in an ATP-dependent reaction catalyzed by glutamine synthetase. • Then the side chain amino group of Gln is further transferred to a-ketoglutarate to form Glu in a reaction catalyzed by glutamate synthase, an enzyme only present in bacteria and plants, not in animals. • The amide group of Gln is a source of nitrogen in the synthesis of a variety of compounds, such as carbamoyl phosphate, Trp, His, glucosamine-6-P, CTP, and AMP. • The amino groups of most other amino acids are derived from glutamate via transamination. Newly fixed nitrogen in the form of NH4+ is first incorporated into glutamate to form glutamine The side chain amino group of glutamine is then e transferred to a-ketoglutarate to form Glu 5. The bacterial Glutamine synthetase is a central control point in nitrogen metabolism • The bacterial glutamine synthetase has 12 identical subunits (each having an independent active site) arranged as two hexagonal rings. • Each subunit of the enzyme is accumulatively inhibited by at least eight allosteric effectors. • In addition the enzyme is more susceptible to the allosteric inhibition by having Tyr397 residue modified by adenylylation. • The addition and removal of the AMP group to the glutamine synthetase are catalyzed by two active sites of the same bifunctional adenylyltransferase (AT). • The substrate specificity of AT was found to be controlled by a regulatory protein, PII. • The activity of PII, in turn, is regulated by the uridylylation of a specific Tyr residue: PII-UMP stimulates the adenylylation activity of AT, however, the unmodified PII stimulates the deadenylylation activity of AT. • The addition and removal of UMP to PII, in turn, are again catalyzed by two active sites of the same protein, uridylyltransferase (UT): a-ketoglutarate and ATP stimulate the uridylylation, however, Gln and Pi stimulate the deuridylylation (thus adenylylation of AT, inactivating glutamine synthetase). The bacterial glutamine synthetase has 12 subunits arranged as two rings of hexamers Active sites Tyr397 (adenylylation site) The glutamine synthetase is accumulatively inhibited by at least 8 allosteric effectors, mostly end products of glutamine metabolism A specific Tyr residue in bacterial glutamine synthetase can be reversibly adenylylated 6. Amidotransferases catalyze the transfer of the amide amino group from Gln to other compounds • All of the enzymes contain two structural domains: one binding Gln, the other binding the amino group accepting substrate. • A Cys residue is believed to act as a nucleophile to cleave the amide bond, forming a covalent glutamylenzyme intermediate with the NH3 produced remain in the active site and react with the second substrate to form an aminated product. A proposed action mechanism for amidotransferases 7. The 20 amino acids are synthesized from intermediates of glycolysis, the citric acid cycle, or pentose phosphate pathway • The nitrogen is provided by Glu and Gln. • As in amino acid degradation, some of the synthetic pathways (e.g., for Ala, Glu, and Asp) are very simple, but others (e.g., the aromatic ones) are very complex. • Most bacteria and plants can synthesize all 20, but mammals can only synthesize about 10, with 10 being “essential”, needed in the diet. • The pathways for the biosynthesis of the 20 amino acids can be categorized into six families according to the metabolic precursors used. + ATP Gln Intermediates of glycolysis, the citric acid cycle, and pentose phosphate pathway serve as precursors for synthesizing the 20 standard amino acids. + acetyl-CoA + pyruvate +Asp 8. a-ketoglutarate is the common precursor for the biosynthesis of Glu, Gln, Pro, and Arg in bacteria • To make Glu, the amino group can be from the side chain of Gln (catalyzed by an amidotransferase, glutamate synthase in bacteria) or ammonia (catalyzed by glutamate dehydrogenase in vertebrates). • Gln is made from Glu by obtaining an amino group from ammonia in a reaction catalyzed by glutamine synthetase. • Proline is synthesized from Glu via one step of phosphorylation (activation), two steps of enzymecatalyzed reductions (using NADPH) and one step of nonenzymatic cyclization reaction (forming a Schiff base). • To make Arg, ornithine is first made from Glu with a few steps similar to that of Pro synthesis, except that an acetyl group is first added to the a-amino group of glutamate to protect the spontaneous Schiff base formation between the aldehyde group and the amino group and removed after an amino group is transferred to the aldehyde group. • Arg is then synthesized from ornithine using steps similar to those of urea cycle in bacteria. • Arg from the diet can be converted to Pro in mammals by being converted to ornithine first using the urea cycle enzymes and then to 1-pyrroline-5carboxylate by ornithine d-aminotransferase. • Similarly, Arg can be formed from glutamate gsemialdehyde (an intermediate of Pro synthesis) also by using ornithine d-aminotransferase. Glu is synthesized from a-ketoglutarate by an amination reaction with amino groups obtained from Gln or ammonia Gln is synthesized from Glu by obtaining an amino group from ammonia in a reaction catalyzed by glutamine synthetase Glutamine synthetase Glutamamate kinase Acetylglutamate synthase Glutamate dehydrogenase spontaneous Pyrroline carboxylate reductase aminotransferase 9. Ser, Gly, and Cys are all derived from 3-phosphoglycerate • Ser is synthesized (in all organisms) from 3phosphoglycerate via NAD+-dependent oxidation, PLP-dependent transamination, and dephosphorylation reactions. • Gly is either derived from Ser by removal of the side chain b-carbon (catalyzed by serine hydroxymethyltransferase, using PLP and H4 folate) or synthesized (in vertebrate liver cells) from CO2 and NH4+ (catalyzed by glycine synthase, using N5,N10-methylene H4 folate). • In plants and bacteria, Cys is made from Ser by replacing the side chain –OH group (after being activated first by acetylation) by an –SH group using sulfide (硫化物) reduced from sulfate (硫酸盐). • In mammals, Cys is made from Met (providing the sulfur atom after being converted to homocysteine) and Ser (providing the carbon skeleton). Ser is synthesized from 3-phosphoglycerate via oxidation, transamination, and dephosphorylation reactions Phosphoserine aminotransferase Gly is either derived from Ser by removal of the side chain b-carbon or synthesized (in vertebrate liver cells) from CO2 and NH4+ Serine hydroxymethyl transferase Glycine Synthase Cys is derived from Ser via a two-step pathway using sulfide in plants and bacteria Cys is synthesized from homocysteine (which is derived from Met) and Ser in mammals Homocysteine (高半胱氨酸) (胱硫醚b-合成酶) (胱硫醚g-裂解酶) Homocysteine is derived from Met via S-adenosylmethionine and Sadenosylhomocysteine intermediates Methionine adenosyl transferase Met ATP Methionine synthase Methyl transferase Hydrolase Homocysteine 10. Ala, Val, and Leu are derived from pyruvate • Ala is synthesized from pyruvate via a simple transamination reaction (with amino group donated by Glu). • Val is synthesized from the transferring of a twocarbon unit to a pyruvate, forming a-acetolactate, which is then converted via isomerization, reduction, dehydration, and transamination. • The immediate precursor of Val, a-ketoisovalerate, is converted to Leu via four steps of reactions: the addition of an acetyl group, a position switching of an –OH group, an oxidative decarboxylation, and a transamination reaction. Val is synthesized from two pyruvates Dihydroxy acid dehydratase Acetohydroxy acid synthase Valine aminotransferase Acetohydroxyl acid isomeroreductase Val Leu is synthesized from a-keto-isovalerate Dehydrogenase Synthase Aminotransferase Isomerase Leu 11. Asp, Asn, Thr, Met are all derived from oxaloacetate • Asp is synthesized from oxaloacetate via a simple transamination reaction. • Asn is synthesized from Asp by an amidation (酰胺 化) reaction with Gln donating the NH4+(catalyzed by an amidotransferase). • Thr and Met are all derived from Asp, branching off after Asp is converted to aspartate b-semialdehyde (in a way similar to Glu to glutamate gsemialdehyde conversion). • Thr is synthesized from aspartate bsemialdehyde via homoserine. • Met is also synthesized from homoserine: it is first activated via succinylation; the accepting the sulfur atom from cysteine and methyl group from N5-methyl H4 folate. Asp is synthesized from oxalocaetate by a simple transamination Transaminase Asn is synthesized from Asp by transferring an amino group from the amide group of Gln Asparagine synthetase Thr is synthesized from Asp via homoserine Threonine synthase Homoserine kinase Homoserine Asp is first dehydrogenase converted to aspartate-b-semialdehyde which is then used to synthesize Thr, Met, and Lys. Met is also synthesized from homoserine via cystathionine and homocysteine Homoserine acyltransferase Cystathionineg-synthase Cystathionineb-lyase Methionine syntase Met 12. Lys and Ile are derived from oxaloacetate and pyruvate • Lys is synthesized from aspartate b-semialdehyde (which is derived from Asp) and pyruvate (contributing to the two carbons at positions of d and e of Lys), with glutamate donating the e-amino group. • Ile is made from a-ketobutyrate (the dehydration/deamination product of Thr) and pyruvate, using the same set of enzymes as that for Val synthesis. Lys is synthesized from aspartate-b-semialdehyde and pyruvate Dihydropicolinate synthase Dihydropicolinate synthase Dehydrogenase Obtaining the e-amino group from Glu Synthase Aminotransferase Removing the carboxyl group of the condensed pyruvate Desuccinylase Epimerase Decarboxylase Lys Pyruvate Thr Acetolactate synthase Acetolactate syntase Ile is made from Thr and Pyruvate using the same set Of enzymes for Val synthesis Acetohydroxy acid isomeroreductase Acetohydroxy acid isomeroreductase a-isopropylmalate synthase Isopropylmalate isomerase Dihydroxyl acid dehydratase Dehydrogenase Valine aminotransferase Leucine aminotransferase Ile Val Leu 13. The aromatic amino acids are derived from phophoenoylpyruvate and erythrose 4-phosphate • All aromatic amino acids share the first seven reactions for biosynthesis, up to the formation of chorismate (分支酸). • All the carbons of the aromatic ring are derived from PEP and erythrose 4-P: ring closure follows the condensation reaction; which in turn is followed by step-wise double bond introduction (by dehydration). • A second PEP enters to make chorismate. • Chorismate can be converted to anthranilate (by accepting an amino group from Gln and releasing a pyruvate) or prephenate (by switching the position of the PEP component via a Claisen rearrangement) by the catalysis of anthranilate synthase or chorismate mutase respectively. • Anthranilate (邻氨基苯甲酸 ) can be further converted to Trp via another four steps of reaction: accepting two carbons from 5-phosphoribosyl-1pyrophosphate (PRPP), and three carbons from Ser. • Prephenate (预苯酸) can be converted to Phe by releasing the carboxyl and hydroxyl groups from the ring, and accepting an amino group from Glu via a transamination reaction. • Prephenate can also be converted to Tyr by an oxidative decarboxylation and a transamination reaction. The biosynthesis of all aromatic amino acids share the first seven steps of reactions four steps three steps one step two steps one step two steps four steps Chorismate, formed from the condensed product of PEP and erythrose 4-P, is the common precursor of Trp, Tyr and Phe. The 2nd PEP enter here Ring formation step 2nd double bond 1st double bond Chorismate Isomerase Synthase Synthase (邻氨基苯甲酸 ) Transferase Trp Synthase Chorismate is converted to Trp by accepting an amino group from Gln, two carbons from 5 phosphoribosyl1-pyrophosphate (PRPP), and three carbons from Ser. Chorismate mutase Dehydrogenase Claison rearrangement dehydratase Chorismate can also be converted to prephenate which serve as the common precursor of Phe and Tyr biosynthesis 14. Tryptophan synthase catalyzes the conversion of indole-3-glycerol phosphate to Trp with indole as an intermediate • The tetrameric enzyme has two a and two b subunits. • Indole-3-glycerol phosphate enters the a subunits (the “lyase”) and is cleaved to form indole and glyceraldehyde-3-P. • Serine enters the b subunits, forms a Schiff base with PLP, and is dehydrated to form a PLPaminoacrylate adduct. • The indole intermediate formed on the a subunit then enters the active site of the b subunit via a “substrate channel”, where it condenses with the PLP-aminoacylate intermediate to form a ketimine (酮亚胺) intermediate, which is then converted to a aldimine (醛亚胺) intermediate. • The aldimine intermediate is then hydrolyzed to form Trp. The a and b subunits of tryptophan synthase have different enzymatic activities with substrate channeling A ketimine An aldimine The hydrophobic indole is channeled from the a to the b subunits in tryptophan synthase 15. Histidine is derived from three precursors: 5-phosphoribosyl 1pyrophosphate, ATP and Gln • The synthesis begins with the condensation of ATP and PRPP: N-1of the purine ring is linked to the C-1 of the ribose. • For the final synthesis of Histidine, PRPP contributes five carbons, ATP contributes one nitrogen and one carbon (both from the purine ring), Gln contributes one nitrogen, and Glu contributes the 3rd nitrogen (of the a-amino group). • The unusual synthesis of histidine from a purine is taken as evidence supporting the hypothesis that life originated from RNA: the synthetic pathway is considered as a “fossil” of the transition from RNA to the more efficient protein-based life forms. Transferase Hydrolase The synthesis of histidine begins with the condensation of PRPP and ATP: The C-1 of the ribose ring is linked to the N-1 of the purine ring. A purine! Hydrolase Cleaved between C-2 and N-3 Amidotransferase Ring opens between N-1 and C-6 Isomerase Aldose Ketose Dehydratase Histidine is made from PRPP, ATP, Gln and Glu. Aminotransferase Dehydrogenase 16. Several feedback inhibition mechanisms are found to work in regulating amino acid biosynthesis • The first case of allosteric feedback inhibition was discovered in studying Ile biosynthesis in E. coli • The enzyme catalyzing the first reaction (threonine dehydratase) is inhibited by the end product (Ile) in a synthetic pathway. • Several intricate feedback inhibition mechansims have been found in branched pathways for amino acid biosyntheses. • In enzyme multiplicity, several isozymes are present to catalyze a common step of reaction: each isozyme responds to a different allosteric modulator, avoiding the inhibition of a common reaction by only one end product. • In concerted inhibition, one enzyme is inhibited by two or more than two modulators, with effect more than additive. • In sequential feedback inhibition, the end products inhibit enzymes catalyzing the branching points only, while the initial committing step is inhibited by common precursors of the end products. The first case of allosteric feedback inhibition was discovered in studying Ile synthesis in E. coli In branching pathways leading to the synthesis of multiple end products from common precursors, several forms of feedback inhibition mechanisms are used to coordinately regulate the synthesis of all the amino acids. Several intricate feedback inhibition mechansims have been found in branched pathways for amino acid biosyntheses Sequential Enzyme multiplicity Concerted High level of one (e.g., Y) should not prevent the synthesis of the other (e.g., Z).