* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download Introduction to Molecular Markers and their
Therapeutic gene modulation wikipedia , lookup
Pharmacogenomics wikipedia , lookup
Genetic engineering wikipedia , lookup
Hardy–Weinberg principle wikipedia , lookup
Designer baby wikipedia , lookup
Genome evolution wikipedia , lookup
Public health genomics wikipedia , lookup
Bisulfite sequencing wikipedia , lookup
Genome (book) wikipedia , lookup
Frameshift mutation wikipedia , lookup
Human genetic variation wikipedia , lookup
Cell-free fetal DNA wikipedia , lookup
Genomic library wikipedia , lookup
Cre-Lox recombination wikipedia , lookup
Genealogical DNA test wikipedia , lookup
No-SCAR (Scarless Cas9 Assisted Recombineering) Genome Editing wikipedia , lookup
Molecular cloning wikipedia , lookup
Genome editing wikipedia , lookup
Site-specific recombinase technology wikipedia , lookup
History of genetic engineering wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Point mutation wikipedia , lookup
Molecular Inversion Probe wikipedia , lookup
Quantitative trait locus wikipedia , lookup
Genetic drift wikipedia , lookup
Population genetics wikipedia , lookup
Polymorphism (biology) wikipedia , lookup
SNP genotyping wikipedia , lookup
Microsatellite wikipedia , lookup
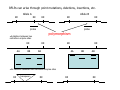
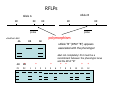
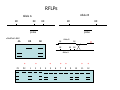
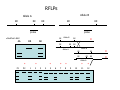
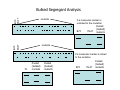
Introduction to Molecular Markers and their Application in Biotechnology Molecular Markers 1. What are molecular markers? – Introduction to technology – Description of types of markers 2. Uses of molecular markers – Application as a genetic tool for plant genotyping and gene mapping – Application in agriculture •Marker-assisted breeding •Plant variety protection •Assessing genetic diversity – Applications in Human Health •Association with genetic-based diseases •Forensic studies Molecular Markers 1. What are molecular markers? – Heritable DNA sequence differences (polymorphisms) – Phenotypically neutral, developmentally and environmentally stable – Identified by techniques such as Southern hybridizations or PCR 2. Types of Markers –Those detected by Southern Hybridizations •RFLPs --Restriction Fragment Length Polymorphisms –VNTRs -- variable number of tandem repeats (minisatellites) –Those detected by PCR-based methods •RAPD -- randomly amplified polymorphic DNA •AFLP -- amplification fragment length polymorphism •CAPS -- cleaved amplified polymorphic site •SSR -- simple sequence repeats (microsatellites) •SNP -- single nucleotide polymorphisms The best molecular markers are those that distinguish multiple alleles per locus (i.e. are highly polymorphic) and are co-dominant (each allele can be observed) •RFLP- a site in a genome where the distance between two restriction sites varies among different individuals. These sites are identified by restriction enzyme digests of chromosomal DNA, and the use of Southern blotting to identify the specific fragments. Requires a radioactive probe! •CAPS-a site in the genome polymorphic for the presence or absence of a restriction enzyme site. Detected by PCR and restriction enzyme digests on gels. •SSR-a site in the genome that contains many short tandem repeat sequences (microsatellites). These sites are usually in the size range of 100-500 base pairs composed of dinucleotide and trinucleotide repeats. They are very polymorphic, scattered through out genomes. Genomes typically contain 1,000s of SSRs! They are detected by PCR using primers flanking the repeats and resolved on gels. •SNP-a single nucleotide difference in the sequence of a gene or segment of the genome. There are typically tens of 1,000s of SNPs and a variety of methods for analyzing them, including highly automated/high throughput procedures with simultaneous scoring of many markers. Detection of SNPs can be done without gels. RFLPs can arise through point mutations, deletions, insertions, etc. RE Allele A RE RE Allele B RE probe probe polymorphism •A deletion between two restriction enzyme sites RE AA BB RE RE RE AB RE AA BB AB •An insertion between two restriction enzyme sites RE transposon RE RE RE RFLPs Restriction Fragment Length Polymorphism (RFLP) probe binding site CHR 1: 5Õ EcoRI ↓ EcoRI 3Õ NNNG|AATTCNNN-------NNNG|AATTCNNN NNNCTTAA|GNNN-------NNNCTTAA|GNNN 3Õ 5Õ probe binding site CHR 2: 5Õ EcoRI ↓ EcoRI 3Õ NNNG|AATTCNNN-------------------NNNG|AATTCNNN NNNCTTAA|GNNN-------------------NNNCTTAA|GNNN 3Õ 5Õ allele A allele B allele C Genomic DNA cut by EcoRI, DNA run on gel, blotted, and hybridized to probe Polymorphism: fragments of different lengths migrate differently in the gel RFLPs Allele B Allele A RE RE RE RE RE probe probe polymorphism •Southern blot AA BB AB •Allele “B” (RFLP “B”) appears associated with the phenotype! •But not completely! #12 must be a recombinant between the phenotypic locus and the RFLP “B” AB BB * P1 P2 * 1 2 * 3 4 5 6 * * 7 8 9 10 * * 11 12 RFLPs Allele B Allele A RE RE RE RE RE probe probe •Southern blot AA BB RE AB Allele B RE * RE RE RE Allele A * P1 P2 * 1 2 * 3 4 5 6 * * 7 8 9 10 * * 11 12 Review of Genetic Linkage and Recombination Genes that are close together on a chromosome tend to be inherited together. In the example shown at left, genes A and B would tend to be inherited together much more often than with gene C. Gene C would be inherited with B slightly more often than with A. Recombination & Crossing Over • Exchange of alleles, but not gene order, between the two homologous chromosomes in diploids during meiosis. • Frequency of crossing-over increases with distance between loci. RFLPs Allele B Allele A RE RE RE RE RE probe probe •Southern blot RE AA BB Allele B * RE AB RE RE RE Allele B Allele A RE RE RE Allele A * P1 P2 * 1 2 * 3 4 5 6 * * 7 8 9 10 * * 11 12 RE * RE * CAPS (cleaved amplified polymorphic sequence) as a molecular marker. RE Allele A RE RE RE Allele B RE Sequence Specific Primers polymorphism •Amplify by PCR •This procedure is much quicker than doing a Southern hybridization, yet yields the same information!! •Digest with restriction enzyme •Visualize on Agarose Gel AA BB AB SSR: simple sequence repeat (length polymorphism) PCR Primer (CA)n (CA)n+4 variation between strains in number of repeats at a given locus PCR yields products of different size: Simple Sequence Repeat (SSR) or Microsatellite Markers Polymorphism is based on the number of times a simple sequence of DNA, usually 2-3 base pairs, is repeated. The variant alleles are probably generated by “stuttering” of DNA polymerase or repair enzymes during DNA replication of repeated sequences. • Male parent (red square) contains alleles 5 and 2 • Female parent (black circle) contains alleles 6 and 3 • Progeny segregate for alleles 2, 3, 5, and 6 • One daughter is mated to a male containing another allele (4), resulting in offspring heterozygous for alleles 5 and 4. Bottom line: interpretation of data for RFLP markers and microsatellites is the same, even though the data is generated by means of different techniques (Southern blotting for RFLP, PCR for microsatellite) Molecular Markers 1. What are molecular markers? – Introduction to technology – Description of types of markers 2. Uses of molecular markers – Application as a genetic tool for plant genotyping and gene mapping – Application in agriculture •Marker assisted breeding •Plant variety protection •Assessing genetic diversity – Applications in Human Health •Association with genetic-based diseases •Forensic studies Genotyping Progeny in a Standard Genetic Cross An example of a genetic cross wild type X (+/+) X F1 teosinte branched mutant (tb1/tb1) all wild type (+/tb1) Normal : tb1 F2 3 : 1 (+/+ or +/tb1) : (tb1/tb1) “Normals” are either +/+ or +/tb1, but one can not distinguish between the two genotypes because “+” is dominant to the recessive “tb1”! •BUT, a molecular marker can easily distinguish between the genotypes! Prepare DNA from leaves of each type and use molecular marker to determine genotype! •Physical Maps •Molecular markers can be mapped relative to one another •This leads to a “physical map” that illustrates the linkage relationships between physical markers on each of the chromosomes •Now, one can map a new mutation (morphological marker) relative to the molecular markers by following linkage between the mutant phenotype and polymorphisms in molecular markers. Each chromosome is divided into bins These bins are about 20 cM Bins are defined by molecular markers •Lets use molecular markers to find the map location of a newly isolated mutant! Map your mutant by comparing its inheritance relative to the inheritance of mapped molecular markers First, make a mapping population: New mutant: tb1-like. This mutant was found in an EMS screen for new mutants in maize inbred B73 tb tb In a B73 background B1 B2 tb B3 B4 B1 B2 tb B3 B4 F1 x x Mo17 M1 M2 TB M3 M4 M1 M2 TB M3 M4 B1 B2 tb B3 B4 M1 M2 TB M3 M4 Generate Mapping Population F1 F2 B1 B2 tb B3 B4 B1 B2 TB B3 B4 B1 B2 TB B3 B4 B1 B2 tb B3 B4 B1 B2 tb B3 B4 B1 B2 TB B3 B4 B1 B2 TB B3 B4 B1 B2 tb B3 B4 WT 1 WT tb mutant 2 1 This population will segregate 3:1 for wild type and tb-like plants. You will isolate DNA from some number of mutant (tb) plants. Every chromosome transmitted is potentially recombinant. Every F2 mutant can be scored for 0/2, 1/2, or 2/2 recombinant chromosomes. For each molecular marker to be tested for linkage with “tb” mutant, identify a polymorphism between the B73 and MO17. Then score inheritance of the polymorphism. •You look to see if the polymorphism is inherited with (linked to) your mutant. B73 Mo17 tb 1 tb 2 tb 3 tb 4 tb 5 tb 6 tb 7 tb 8 tb 9 tb 10 tb 11 tb 12 tb 13 tb 14 tb 15 tb 16 tb 17 tb 18 tb 19 tb 20 If a molecular marker is very closely linked to the mutation (tb), then those progeny homozygous for the mutation will also be homozygous for the B73 polymorphism B73 Mo17 tb 1 tb 2 tb 3 tb 4 tb 5 tb 6 tb 7 tb 8 tb 9 tb 10 tb 11 tb 12 tb 13 tb 14 tb 15 tb 16 tb 17 tb 18 tb 19 tb 20 If a molecular marker is unlinked to the mutation (tb), then those progeny homozygous for the mutation are just as likely to inherit the MO17 polymorphism as the B73 polymorphism •20 recombinant chromosomes out of a total of 40 scored •This marker is unlinked to the mutation! B73 Mo17 tb 1 tb 2 tb 3 tb 4 tb 5 tb 6 tb 7 tb 8 tb 9 tb 10 tb 11 tb 12 tb 13 tb 14 tb 15 tb 16 tb 17 tb 18 tb 19 tb 20 If a molecular marker is linked to the mutation (tb), then those progeny homozygous for the mutation are more likely to inherit the B73 polymorphism then the MO17 polymorphism •8 recombinant chromosomes out of a total of 40 scored 8/40 = 0.2 or 20% recombination or 20 map units (20 cM between this marker and the mutation). •You have found a linked molecular marker!! B1 B2 tb B3 B4 B73 Mo17 tb 1 tb 2 tb 3 tb 4 tb 5 tb 6 tb 7 tb 8 tb 9 tb 10 tb 11 tb 12 tb 13 tb 14 tb 15 tb 16 tb 17 tb 18 tb 19 tb 20 If a molecular marker is linked to the mutation (tb), then those progeny homozygous for the mutation are more likely to inherit the B73 polymorphism then the MO17 polymorphism •8 recombinant chromosomes out of a total of 40 scored 8/40 = 0.2 or 20% recombination or 20 map units (20 cM between this marker and the mutation). •You have found a linked molecular marker!! B1 B2 tb B3 B4 mutants Mo17 B73 B73 Mo17 Bulked Segregant Analysis If a molecular marker is unlinked to the mutation Pooled (bulked) B73 Mo17 mutants mutants If a molecular marker is linked to the mutation F1 Pooled (bulked) normals Pooled (bulked) mutants B73 Mo17 Pooled (bulked) mutants SNP: single nucleotide polymorphisms ¾ Outgrowth of sequencing projects Detection of SNPs can be done without gels: highly automated/high throughput and/or highly parallel (simultaneous scoring of MANY markers) Allele-Specific Extension & Identification in CE: “Minisequencing” (ABI SNaPShotTM) Degree of Multiplexing Depends on Resolution dR6G dR110 ABI SNaPshot® on 3130xl Genotyping by SBE and Mass Spectrometry PCR Amplification SAP Treatment Single Base Extension Spot on 384-place Chips MALDI-TOF Mass Spec Other uses for Molecular Markers •Plant Variety Protection •Verify varietal identity, purity and stability •Marker-Assisted Breeding •For accelerated trait/transgene introgressions •For introgression of quantitative traits •Estimation of Genetic Variation •For phylogenetic analysis •For preservation of rare plant and animal species •For management of wild species •Plant Pathogen Identification Molecular Markers in Human Health •Identification of human genetic diseases •Sickel cell anemia •Huntingtons disease •Tay Sachs disease •Cystic Fibrosis (and many more) •Forensic Science •Paternity Determinations Introduction to Molecular Markers and their Application in Biotechnology