* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download 350-Cell Cycle-DF - Department Of Biological Sciences Hunter
Survey
Document related concepts
Signal transduction wikipedia , lookup
Extracellular matrix wikipedia , lookup
Tissue engineering wikipedia , lookup
Cytokinesis wikipedia , lookup
Cell encapsulation wikipedia , lookup
Cell growth wikipedia , lookup
Organ-on-a-chip wikipedia , lookup
Cellular differentiation wikipedia , lookup
Cell culture wikipedia , lookup
Biochemical switches in the cell cycle wikipedia , lookup
Programmed cell death wikipedia , lookup
Transcript
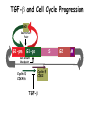
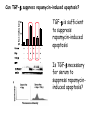
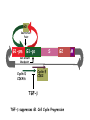
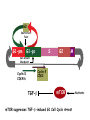
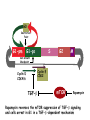
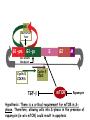
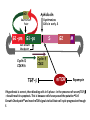
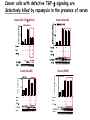
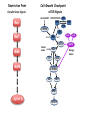
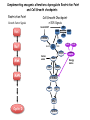
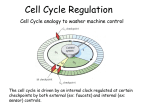
TGF- and Cell Cycle Progression G0 Restriction Point G1-pm G1-ps S Cell Growth Checkpoint Cyclin D CDK4/6 Cyclin E CDK2 TGF- G2 M Effect of rapamycin on cell cycle progression in MDA-MB-231 cells G1 S G2/M G1 S G2/M G1 S G2/M Sub genomic Rapamycin induces primarily G1 arrest in the presence of serum - and apoptosis in the absence of serum Can TGF- suppress rapamycin-induced apoptosis? TGF- is sufficient to suppress rapamycin-induced apoptosis Is TGF- necessary for serum to suppress rapamycininduced apoptosis? Is TGF- in serum necessary for serum to suppress rapamycin-induced apoptosis % Non-Viable Cells Figure 3A 100 % Non-Viable 50 Cells Series1 0 1 - 2 - + 3 + Rap - + + + TGF- -Ab - - - + Serum 4 Cl PARP actin TGF- is necessary for serum to suppress rapamycin-induced apoptosis Summary: •Rapamycin induces apoptosis in MDA-MB-231 cells in the absence of serum •In the presence of serum, rapamycin induces G1 arrest •TGF- is sufficient to suppress rapamycin-induced apoptosis in the absence of serum •TGF- present in serum is necessary for serum to suppress rapamycin-induced apoptosis Question: Why does rapamycin induce apoptosis when TGF- is absent? G0 Restriction Point G1-pm G1-ps S G2 Cell Growth Checkpoint Cyclin D CDK4/6 Cyclin E CDK2 TGF- TGF- suppresses G1 Cell Cycle Progression M G0 Restriction Point G1-pm G1-ps S G2 M Cell Growth Checkpoint Cyclin E CDK2 Cyclin D CDK4/6 TGF- mTOR mTOR suppresses TGF--induced G1 Cell Cycle Arrest Nutrients G0 Restriction Point G1-pm G1-ps S G2 M Cell Growth Checkpoint Cyclin E CDK2 Cyclin D CDK4/6 TGF- mTOR Rapamycin Rapamycin reverses the mTOR suppression of TGF- signaling and cells arrest in G1 in a TGF--dependent mechanism G0 Restriction Point G1-pm G1-ps S G2 M Cell Growth Checkpoint Cyclin D CDK4/6 Cyclin E CDK2 X TGF- mTOR Rapamycin If TGF- signaling is suppressed or defective, there is no G1 arrest with rapamycin treatment - and now the cells die in the presence of rapamycin - Why? G0 Restriction Point G1-pm G1-ps S G2 M Cell Growth Checkpoint Cyclin E CDK2 Cyclin D CDK4/6 TGF- mTOR Rapamycin Hypothesis: There is a critical requirement for mTOR in Sphase. Therefore, allowing cells into S-phase in the presence of rapamycin (ie w/o mTOR) could result in apoptosis G0 Aphidicolin Restriction Point G1-pm Synchronizes Cells in early S G1-ps S G2 M Cell Growth Checkpoint Cyclin E CDK2 Cyclin D CDK4/6 TGF- mTOR Rapamycin If hypothesis is correct, then blocking cells in S-phase - in the presence of serum/TGF- - should result in apoptosis. This is because cells have passed the putative “Cell Growth Checkpoint” and need mTOR signals to facilitate cell cycle progression through S Blocking cells in S-phase with aphidicolin sensitizes cells to rapamycin In the presence of serum/TGF- - if cells are allowed to enter S-phase, then the lack of mTORC1 signals to 4E-BP1 tells the cell that nutrients are in short supply and that replicating the genome is probably a bad career move! % Non-Viable Cells Figure 6A 100 % Non-Viable 50 Cells Series1 0 Rap Aph Cl PARP actin 1 - 2 + 3 - 4 - + + + The cells then do the honorable thing – and commit suicide IMPLICATION: Cancer cells with defective TGF- signaling could be selectively killed by rapamycin in the presence of either serum or TGF- Importantly: Many cancers have defects in TGF- signaling – especially Smad4 - that is critical for suppression of G1 cell cycle progression Cancer cells with defective TGF- signaling are Selectively killed by rapamycin in the presence of serum Breast (Smad4) Breast (No TGF- defect) MDA-MB-231 Breast (PKCδ) 100 SW480 50 Series1 % Non Viable Cells % Non Viable Cells Colon (Smad4) 100 BT-549 50 Series1 0 0 Serum + - - + - Serum + - - + - Rap - - + + + Rap - - + + + TGF- - - - - + TGF- - - - - + Cl PARP actin Cl PARP actin Summary: 1) If TGF- is present, rapamycin induces cell cycle arrest in G1 - by increasing TGF- signaling 2) In the absence of TGF- signaling, rapamycin does not arrest cells in late G1 and they progress through the remainder of G1 into S-phase 3) However, if cells progress into S-phase in the presence of rapamycin, they undergo apoptosis rather than arrest - because of an apparent stringent requirement for mTOR during S-phase Cell Growth Checkpoint Rapamycin induces arrest Rapamycin induces apoptosis G1 S Nutrients Cyclin D-CDK4/6 Cyclin E-CDK2 p27 Rapamycin PLD mTOR TGF- Survival Signals PI3K Growth Factors Implications: In addition to the rapamycin concentration issues – another complication is that in most cancers, rapamycin is cytostatic rather than cytotoxic Therefore - Defective TGF- signaling may be an Achilles heel for strategies that target mTOR - especially in colon and pancreatic cancers where defects in TGF- signaling are common Defective TGF- signaling creates a “Synthetic Lethality” for strategies that suppress the phosphorylation of 4E-BP1 by mTORC1 Alternatively Strategies that suppress mTOR could be combined with strategies that suppress TGF- signaling – creating a synthetic lethal situation Complementary Signals promote G1 Cell Cycle Progression G0 (Quiescence) Restriction Point G1-pm G1-ps Cell Growth Checkpoint (mTOR) Growth Factor Signals Tyrosine kinases Ras/Raf/MEK/MAPK S G2 Gatekeepers Myc SV40 Early Region (Suppression of p53, Rb and PP2A) M Growth Factor Signals Restriction Point G1-pm G1-ps Nutritional Sufficiency Cyclin D-CDK4/6 Amino acids Fatty acids Energy ATP RalA O2 Vps34 Cell Size Nutritional Sufficiency Cell Growth Checkpoint (START) S Cyclin E-CDK2 G0 Cyclin A-CDK2 Rheb PLD mTOR TGF- Commitment Cell Growth Conventional View of Cell Cycle Points: The Restriction Point, originally characterized by Arthur Pardee, is a point in G1 where cells no longer require growth factors and commit to completing the cell cycle In the absence of growth factors, cells exit the cell cycle into quiescence or G0 Zetterberg and colleagues have mapped the Restriction Point to a site ~ 3.5 hr after mitosis - where cyclin D is elevated Figure 8.8 The Biology of Cancer (© Garland Science 2007) From: Weinberg, The Biology of Cancer, 2007 Leland Hartwell described a site in the Yeast cell cycle called START that is late in G1 where cells evaluate whether there is sufficient nutrition to complete cell division In some texts, the Restricition Point is referred to as the mammalian equivalent of START - and located near the site where cyclin E is activated Rapamycin treatment results in the activation of TGF- signaling and arrest at the cyclin E site - that can be clearly distinguished both temporally and genetically from the growth factor-dependent Restriction Point Genetic requirements for the transformation of human cells (I) (Hahn et al., Nature 400:464, 1999; MCB 22;2111, 2002) Genetic effect Molecular Target Cell cycle target Ras Growth factor signals Restriction point SV40 Large T p53 Rb G1/S checkpoint All G1 checkpoints SV40 small t PP2A Cell Growth checkpoint (?) Genetic requirements for the transformation of human cells (II) (Boehm et al., MCB 25:6464, 2005) Genetic effect Molecular Target Cell cycle target Ras Growth factor signals Restriction point p53 KO Rb KO p53 Rb G1/S checkpoint All G1 checkpoints Myc PTEN KO Gene expression mTORC1 Cell Growth checkpoint (?) Cell Growth checkpoint (?) Restriction Point Cell Growth Checkpoint Growth Factor Signals Ras mTOR Signals PI3K Insulin/IGF1 PIP3 PIP2 PTEN mTORC2 PDK1 Raf Ser473 Akt T308 TSC1/2 Mek Amino acids Rheb PLD1 MAPK FKBP38 TGF- Myc Cyclin D LKB1 AMPK Energy status mTORC1 S6K AMP Cyclin E Growth factor, essential amino acid, glutamine, and suppression of mTOR block G1 cells cycle progression at distinguishable sites in G1 A temporal relationship can be established whereby the GFdependent R is upstream from sites that are sensitive to EAA, Q, and mTOR suppression Temporal mapping of G1 cell cycle checkpoints Cells require 12- 16 hr to enter S-phase after GF deprivation or mTOR suppression Cells require 20+ hr to enter S-phase after amino acid deprivation For cells arrested in G0 by GF deprivation, amino acid deprivation blocks entry into Sphase for 12-14 hr For cells arrested in G0 by GF deprivation, mTOR suppression blocks entry into Sphase for 16-18 hr Blocking G1 cell cycle progression by growth factor and nutrient deprivation, and suppression of mTOR A B P-Rb S807/811 D Cyclin D CM (hrs.) P-AktT308 -GF P-AktS473 -EAA P-S6KT389 P-4EBP1 LC3-II p85 p70 D2/D3 D1 -Q +Rapa. C Akt Pan-Rb E p21 CM (hrs.) S6K -GF 4EBP1 -EAA Actin -Q +Rapa. Different blocking conditions have differential effects on cell cycle regulators and on autophagy – notably for EAA and Q Summary Data supports a model where there is GF-dependent R where multi-cellular organisms determine whether it is appropriate for a cell to divide During G1-ps, cells that have been given the green light to divide, determine whether they have the means/raw materials to double the mass of a cell, Replicate its genome, and divide into two daughter cells The late G1 “Metabolic Checkpoints” in late G1 collectively represent a “Cell Growth” checkpoint that responds to nutrients that is evolutionarily equivalent to START in the yeast cell cycle TOR/mTOR is likely the ultimate arbiter for determining nutrient sufficiency Complementing oncogenic alterations dysregulate Restriction Point and Cell Growth checkpoints Restriction Point Cell Growth Checkpoint Growth Factor Signals mTOR Signals Ras PI3K Insulin/IGF1 PIP3 PIP2 PTEN mTORC2 PDK1 Raf Ser473 Akt T308 TSC1/2 Mek Amino acids Rheb PLD1 MAPK FKBP38 TGF- Myc Cyclin D LKB1 AMPK Energy status mTORC1 S6K AMP Cyclin E Dysregulated metabolic checkpoints in cancer cells In response to amino acid deprivation, MDA-MB-231 breast and Panc1 pancreatic cells arrest in S-phase and G2/M Conclusions The GF-dependent R can be distinguished from late G1 metabolic checkpoints and mTOR The G1 metabolic checkpoints – like R – are dysregulated in human cancer cells Cooperating genetic alterations in cancer cells disable both R and the late metabolic checkpoints that collectively may represent a “Cell Growth” checkpoint with mTOR as the final arbitor Surprisingly, mTOR, which is widely known to be regulated by amino acids, blocked cell cycle progression well downstream of the amino acid sites It is hypothesized that other nutrient inputs – such as glucose and phosphatidic acid (lipids) may be required for complete activation of mTOR and progression into S-phase