* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download Mutation Rates
Mitochondrial DNA wikipedia , lookup
Cancer epigenetics wikipedia , lookup
Zinc finger nuclease wikipedia , lookup
DNA damage theory of aging wikipedia , lookup
Skewed X-inactivation wikipedia , lookup
Genetic drift wikipedia , lookup
Genome (book) wikipedia , lookup
Site-specific recombinase technology wikipedia , lookup
Gene therapy of the human retina wikipedia , lookup
Cell-free fetal DNA wikipedia , lookup
Saethre–Chotzen syndrome wikipedia , lookup
No-SCAR (Scarless Cas9 Assisted Recombineering) Genome Editing wikipedia , lookup
Genome evolution wikipedia , lookup
Koinophilia wikipedia , lookup
BRCA mutation wikipedia , lookup
Oncogenomics wikipedia , lookup
Microsatellite wikipedia , lookup
Population genetics wikipedia , lookup
Microevolution wikipedia , lookup
Haplogroup G-P303 wikipedia , lookup
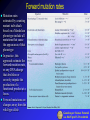
Mutation Rates Ultimately, the source of genetic variation observed among individuals in populations is gene mutation. Mutation generates new alleles, and these are the substance of all evolutionary change. The mutation rate is defined as the probability that a copy of an allele changes to some other allelic form in one generation. Mutation rates at the gene level depends on mutation rates at other levels: Mutation rates for different kinds of mutations can be expressed as mutations per locus, per gene, per nucleotide, and per gamete. All of these indicate a specific type of mutation occurring per generation (higher eucaryotes) or per DNA replication (microorganisms), reflecting mutations arising anew in the unit time. In addition, mutations rates may be expressed is relation to visible phenotypes or in relation to of DNA sequence changes Therefore, it is useful distinguishing between mutation rates: per base pair per generation (or replication) per gene per generation (or replication) per genome or gamete per generation (or replication) Genetica per Scienze Naturali a.a. 04-05 prof S. Presciuttini Estimating the mutation rate in the fluctuation test Culture number Number T1 mutants R Total number of cells 1 1 0.2 x 10 8 2 0 0.2 x 10 8 3 3 0.2 x 108 4 0 0.2 x 10 8 5 0 0.2 x 10 8 6 5 0.2 x 10 8 7 0 0.2 x 10 8 8 5 0.2 x 108 9 0 0.2 x 108 10 6 0.2 x 10 11 107 0.2 x 108 12 0 0.2 x 10 13 0 0.2 x 108 14 0 0.2 x 108 15 1 0.2 x 10 16 0 0.2 x 108 17 0 0.2 x 10 18 64 0.2 x 108 19 0 20 35 0.2 x 108 0.2 x 108 Total 227 4 x 10 8 8 8 8 Luria and Delbrück's fluctuation test provides a way of measuring the mutation rate towards acquisition of resistance to phage T1. Examine in more detail the results of their experiment relative to the 20 small cultures. The total number of mutant cells was 227, out of a total population of 4 x 108 cells. This leads to a frequency of mutant cells in the total population of 5.7 x 10-7. Note that this is the proportion of mutants among all the living cells at the end of the experiment; this is not the mutation rate. 8 8 Mutation frequency = 231 / 4x10 = 5.7 x 10-7 Genetica per Scienze Naturali a.a. 04-05 prof S. Presciuttini Mutation frequency and mutation rate To estimate the mutation rate, we need to estimate how many cell duplications occurred to produce the extant population in each culture, and to estimate how many times a mutation toward resistance occurred among all these cell duplications Difference between mutation frequency and mutation rate. • The number of mutant cells in the final population (M) of this example is 10, and the total number of cells (N) is 16 x 4 = 64 (mutation frequency = M/N =1.6 x 10-1). • The mutation rate m is the number of mutation events (n = 2) among the total number of cell duplications D (= 15 x 4 = 60); thus m = n/D = 3.3 x 10-2. Genetica per Scienze Naturali a.a. 04-05 prof S. Presciuttini Estimating mutation rates in bacteria In the real case, we cannot count the number of independent mutation events the occurred in each culture; we can only distinguish among the cultures in which no mutation had occurred (zero mutants) from all the others. Thus, the 20 cultures that were tested for phage T1 resistance can be divided among those in which no mutation had occurred (11 of the 20 = 0.55) and those in which a mutation had occurred once or more. This situation is well described by the Poisson distribution, in which the probability of zero occurrences of an event (P0) is given by P0 = e-n, where n is the probability that an event will occur. We can estimate n from the data by letting 0.55 = e-n and solving for n (n = -ln 0.55); this gives n = 0.6 mutation events per tube. We can now estimate the number of cell duplications (D) that occurred in each tube. If N is very large, and if the original number of cells was very small, then a sufficiently accurate estimate of D is given by N itself. Thus, since D = 0.2 × 108, we arrive at an estimate of the mutation rate m = 0.6/ 0.2×108 = 3 × 10-8. Note that m is expressed as number of mutations per gene per DNA replication. Genetica per Scienze Naturali a.a. 04-05 prof S. Presciuttini Forward mutation rates Mutation rates estimated by counting mutant individuals based on a Mendelian phenotype include all mutations that cause the appearance of that phenotype In practice, this approach estimate the forward mutation rate, or any DNA change that abolishes or severely impede the production of a functional product at a locus. Forward mutations are changes away from the wild-type allele Genetica per Scienze Naturali a.a. 04-05 prof S. Presciuttini Forward and Backward mutation rates If we consider a locus with two possible alleles (A and a)) then we can consider a forward (u) and backward mutation (v). Forward mutation is the mutation from wildtype allele to the detrimental allele. Backward mutations undo the forward mutation. Because there are many ways to destroy the function but fewer ways to undo that harm, backward mutations are normally more rare than forward mutations. Genetica per Scienze Naturali a.a. 04-05 prof S. Presciuttini Estimated mutation rates Bacteria, Archae, and Eukaryotic microbes produce about one mutation per 300 chromosome replications. For E. coli this works out to be between 10-6 and 10-7 mutations per gene per generation, however it is important to note that there are certain "hot spots" or "cold spots" for spontaneous mutations. Higher eukaryotes have the same rate of spontaneous mutation, so that rates per sexual generation are about one mutation per gamete (close to the maximum compatible with life). RNA viruses have much higher mutation rates - about one mutation per genome per chromosome replication - and even small increases in their mutation rates are lethal. Because a complex individual has a trillion or so nucleotides, each individual is likely to sustain one or more mutations. Rates of expressed gene mutations average about 1 per 100,000 to 1 per million: rates of expression of phenotypic effects are often higher because they are controlled by many genes Genetica per Scienze Naturali a.a. 04-05 prof S. Presciuttini Nucleotide mutation rate Rates of spontaneous mutation seem to be determined by evolutionary balances between the deleterious consequences of too many mutations and the additional energy and time required to further reduce mutation rates. In microorganisms, the rate of mutation for any nucleotide (point mutations) is generally included between 10-9 and 10-10 per DNA replication. Although this rate of mutation may seem exceedingly small, the total amount of new genetic variation introduced by spontaneous mutation at each DNA replication is significant. Consider the genome of E. coli, of the size of about 5 x 106 bp. With a mutation rate intermediate between those listed above (say 5 x 10-10), 25 x 10-4, or one every 400 cells carries a new point mutation. This means that in a single large bacterial culture (1 litre), in which concentrations of 2 x 109 cells/ml are easily obtained (=2 x 1012 total cells), some 5 x 109 new mutations are present, corresponding to 1,000 mutations for each base pair. In practice, all possible nucleotide substitutions and all possible single insertion/deletions, as well as many large rearrangements are represented in a moderately large bacterial population Genetica per Scienze Naturali a.a. 04-05 prof S. Presciuttini Measuring mutation rates in human The mutation rate is a measure of the frequency of a given mutation per generation (or per gamete, which is equivalent). Ordinarily, rates are given for specific loci. Thus the mutation rate for achondroplasia is 6-13 mutants per million gametes. This means that each gamete has about 1 chance in 100,000 of carrying a new mutation for achondroplasia. Mutation rates are based almost exclusively on rare autosomal dominant or X-linked recessive traits. It is virtually impossible to measure autosomal recessive traits accurately. The range of known mutation rates varies from 1 in 10,000 for Duchenne muscular dystrophy and neurofibromatosis type-1 (the largest genes known) to several genes in the range of 1 in 10,000,000. Mutation rate studies never measure all the possible mutations at a locus. Many of the mutations cause no obvious phenotypic effect and could only be recognized by direct analysis of DNA sequences. Genetica per Scienze Naturali a.a. 04-05 prof S. Presciuttini A recent work about mutation rate in human The approach by Nachman and Crowell to measuring the human mutation rate takes advantage of the well-known result that for neutral mutations, the mutation rate is equal to the rate of mutation substitution Knowing divergence time and generation length of two species allowed them to estimate of the rate and pattern of mutation in stretches of DNA without function They sequenced 18 pseudogenes in humans and chimpanzees, including 12 on autosomes and 6 on the X chromosome. In this way, they 1. estimated the average mutation rate per nucleotide site 2. compared mutation rates for different sites and for different classes of mutation to evaluate heterogeneity of mutation rate 3. compared rates of divergence on the X chromosome and on autosomes to evaluate the hypothesis that the X chromosome has a lower mutation rate than the autosomes 4. provided an approximation of the genomic deleterious mutation rate by considering the total mutation rate and the fraction of the genome that is subject to constraint Genetica per Scienze Naturali a.a. 04-05 prof S. Presciuttini Genetica per Scienze Naturali a.a. 04-05 prof S. Presciuttini Nachman and Crowell main results Genetica per Scienze Naturali a.a. 04-05 prof S. Presciuttini Nachman and Crowell results (1) Genetica per Scienze Naturali a.a. 04-05 prof S. Presciuttini Nachman and Crowell results (2) Genetica per Scienze Naturali a.a. 04-05 prof S. Presciuttini Nachman and Crowell results (3) Genetica per Scienze Naturali a.a. 04-05 prof S. Presciuttini Nachman and Crowell results (4) Genetica per Scienze Naturali a.a. 04-05 prof S. Presciuttini Summary of mutation rates in human Mutation rates per generation • Per base pair ~10-8 • Per gene ~10-6 - 10-5 • Per genome All point mutations: ~100 per gamete Deleterious mutations: 1 ~ 2 per gamete The rate of nucleotide substitutions is on the order of 1 per 100,000,000 nucleotides. Since there are 3 billion nucleotides per genome, that means that every gamete has about 30 new mutations involving nucleotide substitutions. Genetica per Scienze Naturali a.a. 04-05 prof S. Presciuttini