* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download IBS Methods for Affected Pairs Linkage
Survey
Document related concepts
Artificial gene synthesis wikipedia , lookup
Skewed X-inactivation wikipedia , lookup
Neuronal ceroid lipofuscinosis wikipedia , lookup
Tay–Sachs disease wikipedia , lookup
X-inactivation wikipedia , lookup
Fetal origins hypothesis wikipedia , lookup
Public health genomics wikipedia , lookup
Population genetics wikipedia , lookup
Neocentromere wikipedia , lookup
Medical genetics wikipedia , lookup
Genome-wide association study wikipedia , lookup
Microevolution wikipedia , lookup
Genetic drift wikipedia , lookup
Human leukocyte antigen wikipedia , lookup
Quantitative trait locus wikipedia , lookup
Transcript
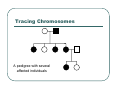
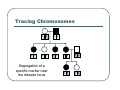
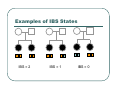
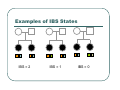
IBS Methods for Affected Pairs Linkage Biostatistics 666 Lecture 14 Genetic Mapping “Compares the inheritance pattern of a trait with the inheritance pattern of chromosomal regions” Positional Cloning “Allows one to find where a gene is, without knowing what it is.” Where are the genes influencing a particular trait? Intuition for Linkage Analysis z Millions of variations could potentially be involved • Costly to investigate each individually z Within families, variation is organized into a limited number of haplotypes • Sample modest number of markers to determine whether each stretch of chromosome is shared Tracing Chromosomes A pedigree with several affected individuals Tracing Chromosomes Segregation pattern for chromosome carrying disease alleles Tracing Chromosomes 1 2 1 3 1 4 Segregation of a specific marker near the disease locus 3 4 2 3 1 3 5 6 3 5 1 5 Tracing Chromosomes 12 12 12 14 14 14 13 13 13 34 34 34 13 14 14 Multiple nearby markers can segregate in a manner that tracks disease! 24 24 24 13 13 13 14 14 14 12 12 12 Today … z Linkage analysis with sibling pairs z Find markers that are near disease locus z Minimalist approach … • Identity-by-State (IBS) based method • Near means recombination fraction θ < ½ Reference for Today… z Power of IBS Methods for Linkage Analysis z Bishop DT and Williamson JA (1990) Am J Hum Genet 46:254-265 z Recommended Reading Bishop and Williamson (1990) Opening Line "The availability of a large number of DNA markers has made possible mapping projects with the certainty that if: (a) a major gene exists for a trait; (b) the trait is reasonably homogeneous; (c) there is sufficient family material available; then a linked marker can be found." Data for a Linkage Study z Pedigree z Observed marker genotypes z Phenotype data for individuals • Set of individuals of known relationship • SNPs, VNTRs, microsatellites Minimalist Approach z Pedigree z Observed Marker Genotypes z Phenotypes • Two individuals of known relationship • A single marker • Both individuals are affected Allele Sharing Analysis z z z Reject random sharing at a particular region Less powerful than classic methods • When disease model is known More robust than classic methods • When disease model is unknown Consider Autosomal Recessive Locus … z For a collection of sibling pairs… z What patterns of sharing do you expect at the disease locus? z What patterns of sharing to you expect as you move away from the disease locus? Historical References z Penrose (1953) suggested comparing IBD distributions for affected siblings. • Possible for highly informative markers (eg. HLA) z Thomson (1986) suggested discarding partially informative families. z Lange (1986) proposed using IBS information instead of IBD. IBS Based Methods z Sample of affected relative pairs z Examine a marker of interest z Count alleles shared for each pair • This includes both … • Chromosomes that are identical-by-descent • Chromosomes that simply carry identical alleles Examples of IBS States 1/ 1 1/1 IBS = 2 1/ 1 1/2 IBS = 1 2/ 2 1/1 IBS = 0 Examples of IBS States 1/ 2 1/2 IBS = 2 1/ 3 1/2 IBS = 1 1/ 2 3/4 IBS = 0 Evidence for Linkage z Increased similarity in affected pairs z Compared to: • Unselected pairs • Unaffected pairs • Discordant pairs • Expectations derived from allele frequencies Test for Independence χ 22df = ∑ i χ12df z z [N IBS =i − E ( N IBS =i )]2 (general test, for sibling pairs) E ( N IBS =i ) 2 2 [ N IBS =0 − E ( N IBS =0 )] [N IBS >0 − E ( N IBS >0 )] = + E ( N IBS =0 ) E ( N IBS >0 ) (grouping often preferable for other relatives) Assuming all counts are relatively large If counts are small, use binomial or trinomial distribution Modeling IBS Sharing z For any relative pair, calculate: • Probability of IBD sharing • 0, 1 or 2 alleles • Conditional probability of IBS sharing • 0, 1, 2 alleles • IBS sharing >= IBD sharing • Why? IBD z The underlying sharing of chromosomes segregating within a family z Siblings share 0, 1 or 2 alleles z • Probabilities ¼, ½ and ¼ Unilineal relatives share 0 or 1 alleles • Probability of sharing is kinship coefficient φ * 4 P(Marker Genotype|IBD State) Relative I II (a,b) (c,d) (a,a) (b,c) (a,a) (b,b) (a,b) (a,c) (a,a) (a,b) (a,b) (a,b) (a,a) (a,a) Prior Probability 0 4papbpcpd 2pa2pbpc pa2pb2 4pa2pbpc 2pa3pb 4pa2pb2 p a4 IBD 1 0 0 0 pa pb p c pa2pb (papb2+pa2pb) pa 3 2 0 0 0 0 0 2papb pa 2 ¼ ½ ¼ Note: Assuming alleles unordered within genotypes P(IBS = i | IBD = j) P( IBS = 2 | IBD = 0) = 2∑ pi2 p 2j + ∑ pi4 i≠ j i P( IBS = 1 | IBD = 0) = 4∑ pi2 p j (1 − pi − p j ) + 4∑ pi3 (1 − pi ) i≠ j i P( IBS = 0 | IBD = 0) = ∑ pi p j (1 − pi − p j ) 2 + ∑ pi2 (1 − pi ) 2 i≠ j i P(IBS = i | IBD = j) P ( IBS = 2 | IBD = 2) = 1 P( IBS = 1 | IBD = 2) = 0 P( IBS = 0 | IBD = 2) = 0 P( IBS = 2 | IBD = 1) = ∑ pi2 i P( IBS = 1 | IBD = 1) = ∑ pi p j i≠ j P( IBS = 0 | IBD = 1) = 0 Example, Assuming Equal Allele Frequencies P(IBS=0) P(IBS=1) P(IBS=2) 2 alleles, IBD=0 .125 .500 .375 2 alleles, IBD=1 .000 .500 .500 3 alleles, IBD=0 .222 .592 .185 3 alleles, IBD=1 .000 .666 .333 Inference from Example z IBS approaches IBD as number of alleles increases z If linkage is being tested with chi-square test, how does the number of alleles (and marker informativeness) affect these two tests: • A test of whether NIBS >= 1 increases? • A test of whether NIBS > 1 increases? IBS Probabilities No. of Alleles P(IBS=0) P(IBS=1) P(IBS=2) 2 .03 .37 .60 3 .05 .48 .47 4 .08 .51 .40 20 .21 .52 .27 ∞ .25 .50 .25 Sibling IBS as a function of allele count, for marker with equally frequent alleles Results of Bishop and Williamson (1990) z Effect size, P(IBS | Affected pair) z Number of alleles at marker z Different relationships z Recombination fraction More Alleles Increase Power Effect of Recombination Varies According to Relationship Power vs. P(IBS | Affected Pair) With no phenocopies, rare alleles are easier to map In general, phenocopies decrease power Shortcomings of IBS Method z All sharing is weighted equally z Inefficient. • Sharing a rare allele • Sharing a common allele • Sharing homozygous genotype • Sharing heterozygous genotype Recommended Reading z Bishop DT and Williamson JA (1990) Am J Hum Genet 46:254-265 z Good introduction to linkage analysis in affected relative pairs, discusses • • • • Marker choice Recombination fraction Disease model Type of relative pair