* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download Tutorial - SigTerms
Saethre–Chotzen syndrome wikipedia , lookup
Epigenetics in learning and memory wikipedia , lookup
Neuronal ceroid lipofuscinosis wikipedia , lookup
X-inactivation wikipedia , lookup
Transposable element wikipedia , lookup
Oncogenomics wikipedia , lookup
Gene therapy of the human retina wikipedia , lookup
Quantitative trait locus wikipedia , lookup
Copy-number variation wikipedia , lookup
Polycomb Group Proteins and Cancer wikipedia , lookup
Genetic engineering wikipedia , lookup
Epigenetics of neurodegenerative diseases wikipedia , lookup
Epigenetics of diabetes Type 2 wikipedia , lookup
Pathogenomics wikipedia , lookup
Essential gene wikipedia , lookup
Public health genomics wikipedia , lookup
Vectors in gene therapy wikipedia , lookup
Gene therapy wikipedia , lookup
History of genetic engineering wikipedia , lookup
Nutriepigenomics wikipedia , lookup
Genomic imprinting wikipedia , lookup
Gene nomenclature wikipedia , lookup
Minimal genome wikipedia , lookup
The Selfish Gene wikipedia , lookup
Therapeutic gene modulation wikipedia , lookup
Ridge (biology) wikipedia , lookup
Gene desert wikipedia , lookup
Genome evolution wikipedia , lookup
Epigenetics of human development wikipedia , lookup
Biology and consumer behaviour wikipedia , lookup
Gene expression programming wikipedia , lookup
Site-specific recombinase technology wikipedia , lookup
Genome (book) wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Microevolution wikipedia , lookup
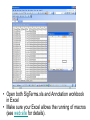
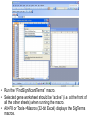
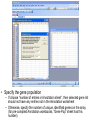
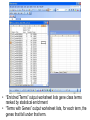
SigTerms tutorial: using the “FindSignificantTerms” macro Chad Creighton, Ph.D. Selecting the Annotation workbook • The SigTerms web site provides links to download pre-compiled Annotation workbooks for several types of gene class associations of potential interest (e.g. GO terms, microRNA targeting predictions). • Annotation workbook should be specific to the gene array that was used for the profiling. – All genes in the workbook should be represented on the array (any genes not on the array should not be represented in the Annotation workbook) – Workbooks specific to particular array platforms are available for download • User can construct his or her own Annotation workbook and specify gene-to-term associations – Details on Annotation workbook format on web site. • Open both SigTerms.xls and Annotation workbook in Excel • Make sure your Excel allows the running of macros (see web site for details). • In the Annotation workbook, insert a new worksheet • (Right-click on the worksheet tab, select “Insert”, and choose “New Worksheet.) • In the new worksheet, paste the list of selected genes in the first column (starting from the first row). • Use Entrez gene number to specify gene – Duplicate Ids are okay. – For statistical calculations, should not include NULL or “---” or “///” containing entries, only entries for genes with Entrez numbers. • Run the “FindSignificantTerms” macro. • Selected gene worksheet should be “active” (i.e. at the front of all the other sheets) when running the macro. • Alt+F8 or Tools->Macros (32-bit Excel) displays the SigTerms macros. • Specify the gene population – If choose “number of entries in Annotation sheet”, then selected gene list should not have any entries not in the Annotation worksheet – Otherwise, specify the number of unique, identified genes on the array (for pre-compiled Annotation workbooks, “Gene Pop” sheet has this number). • “Enriched Terms” output worksheet lists gene class terms ranked by statistical enrichment • “Terms with Genes” output worksheet lists, for each term, the genes that fall under that term. • If desired, one can link additional information to the genes (or microRNAs) listed in the output worksheets. • Can use “MATCH” and “INDEX” Excel functions (Illustrated above and in our Excel tutorial).