* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download Enhancer
Human genetic variation wikipedia , lookup
Cre-Lox recombination wikipedia , lookup
Gene therapy wikipedia , lookup
Short interspersed nuclear elements (SINEs) wikipedia , lookup
Non-coding RNA wikipedia , lookup
Genomic imprinting wikipedia , lookup
Transposable element wikipedia , lookup
Genetic engineering wikipedia , lookup
No-SCAR (Scarless Cas9 Assisted Recombineering) Genome Editing wikipedia , lookup
DNA vaccination wikipedia , lookup
Genome (book) wikipedia , lookup
Epitranscriptome wikipedia , lookup
Ridge (biology) wikipedia , lookup
Public health genomics wikipedia , lookup
Oncogenomics wikipedia , lookup
Gene therapy of the human retina wikipedia , lookup
Transcription factor wikipedia , lookup
Histone acetyltransferase wikipedia , lookup
Gene desert wikipedia , lookup
Point mutation wikipedia , lookup
Human genome wikipedia , lookup
Epigenetics wikipedia , lookup
Epigenetics of depression wikipedia , lookup
Gene expression programming wikipedia , lookup
Genome evolution wikipedia , lookup
Vectors in gene therapy wikipedia , lookup
History of genetic engineering wikipedia , lookup
Helitron (biology) wikipedia , lookup
Epigenetics in stem-cell differentiation wikipedia , lookup
Gene expression profiling wikipedia , lookup
Epigenetics of neurodegenerative diseases wikipedia , lookup
Cancer epigenetics wikipedia , lookup
Microevolution wikipedia , lookup
Designer baby wikipedia , lookup
Non-coding DNA wikipedia , lookup
Site-specific recombinase technology wikipedia , lookup
Long non-coding RNA wikipedia , lookup
Primary transcript wikipedia , lookup
Epigenetics of diabetes Type 2 wikipedia , lookup
Epigenetics in learning and memory wikipedia , lookup
Polycomb Group Proteins and Cancer wikipedia , lookup
Epigenomics wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Epigenetics of human development wikipedia , lookup
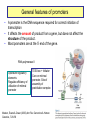
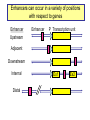
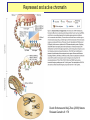
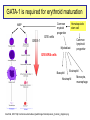
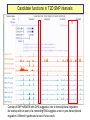
Genomics of Gene Regulation ANSC 497B Ross Hardison Nov. 10, 2009 DNA sequences involved in regulation of gene transcription Protein-DNA interactions Chromatin effects Distinct classes of regulatory regions Act in cis, affecting expression of a gene on the same chromosome. Cis-regulatory modules (CRMs) Maston G, Evans S and Green M (2006) Annu Rev Genomics Hum Genetics 7:29-59 General features of promoters • A promoter is the DNA sequence required for correct initiation of transcription • It affects the amount of product from a gene, but does not affect the structure of the product. • Most promoters are at the 5’ end of the gene. RNA polymerase II Upstream regulatory elements: Regulate efficiency of utilization of minimal promoter TATA box + Initiator: Core or minimal promoter. Site of assembly of preinitiation complex Maston, Evans & Green (2006) Ann Rev Genomics & Human Genetics, 7:29-59 Conventional view of eukaryotic gene promoters Maston, Evans & Green (2006) Ann Rev Genomics & Human Genetics, 7:29-59 Most promoters in mammals are CpG islands TATA, no CpG island About 10% of promoters CpG island, no TATA About 90% of promoters Carninci … Hayashizaki (2006) Nature Genetics 38:626 Fraction of mRNAs Differences in specificity of start sites for transcription for TATA vs CpG island promoters Carninci … Hayashizaki (2006) Nature Genetics 38:626 Enhancers • Cis-acting sequences that cause an increase in expression of a gene • Act independently of position and orientation with respect to the gene. CRM pr luciferase UCE pr lacZ Tested UCE Pennacchio et al., http://enhancer.lbl. gov/ About half of the enhancers predicted by interspecies alignments are validated in erythroid cells Wang et al. (2006) Genome Research 16:1480- 1492 Over half of ultraconserved noncoding sequences are developmental enhancers Pennacchio et al. (2006) Nature 444:499-502 CRMs are clusters of specific binding sites for transcription factors Hardison (2002) on-line textbook Working with Molecular Genetics http://www.bx.psu.edu/~ross/ Enhancers can occur in a variety of positions with respect to genes Enhancer Upstream Enhancer P Transcription unit Adjacent Downstream Internal Distal Ex1 Ex2 Silencer • Cis-acting sequences that cause a decrease in gene expression • Similar to enhancer but has an opposite effect on gene expression • Gene repression - inactive chromatin structure (heterochromatin) • • SIR proteins (Silent Information Regulators) Nucleates assembly of multi-protein complex – hypoacetylated N-terminal tails of histones H3 and H4 – methylated N-terminal tail of H3 (Lys 9) Insulators and boundaries • A boundary in chromatin marks a transition from open to closed chromatin • An insulator blocks activation of promoter by an enhancer – Requires CTCF • Example: HS4 from chick HBB complex has both functions Pr neoR Insulator Enhancer Neo-resistant colonies % of maximum 10 Silencer 50 100 Repression by PcG proteins via chromatin modification Polycomb Group (PcG) Repressor Complex 2: ESC, E(Z), NURF-55, and PcG repressor SU(Z)12 Methylates K27 of Histone H3 via the SET domain of E(Z) me3 K27 H3 N-tail OFF trx group (trxG) proteins activate via chromatin changes • SWI/SNF nucleosome remodeling • Histone H3 and H4 acetylation • Methylation of K4 in histone H3 – Trx in Drosophila, MLL in humans • http://www.igh.cnrs.fr/equip/cavalli/link.PolycombTeaching.html#Part_ 3 Me1,2,3 K4 H3 N-tail ON Histone modifications modulate chromatin structure H3K4me2, 3 http://www.imt.uni-marburg.de/bauer/images/fig2.jpg H3K27me3 Uta-Maria Bauer Repressed and active chromatin Dustin Schones and Keiji Zhao (2008) Nature Reviews Genetics 9: 179 Biochemical features of DNA in CRMs Accessible to cleavage: DNase hypersensitive site Clusters of binding site motifs Bound by specific transcription factors Coactivators Pol Pol IIa II Associated with RNA polymerase and general transcription factors Nucleosomes with histone modifications: Acetylation of H3 and H4 Methylation of H3K4 Lack of methylation at H3K27 or H3K9 … Methods in Genomics of Gene Regulation Chromatin immunoprecipitation: Greatly enrich for DNA occupied by a protein Elaine Mardis (2007) Nature Methods 4: 613-614 ChIP-chip: High throughput mapping of DNA sequences occupied by protein http://www.chiponchip.org Bing Ren’s lab Enrichment of sequence tags reveals function Barbara Wold & Richard M Myers (2008) “Sequence Census Methods” Nature Methods 5:19-21 Illumina (Solexa) short read sequencing - 8 lanes per run - 10 M to 20 M reads of 36 nucleotides (or longer) per run. - 1 lane can produce enough reads to map locations of a transcription factor in a mammalian genome. Example of ChIP-seq ChIP vs NRSF = neuron-restrictive silencing factor Jurkat human lymphoblast line NPAS4 encodes neuronal PAS domain protein 4 Johnson DS, Mortazavi A, Myers RM, Wold B. (2007) Genome-Wide Mapping of in Vivo Protein-DNA Interactions. Science 316:1497-1502. ChIP-seq for chromatin modifications Dustin Schones and Keiji Zhao (2008) Nature Reviews Genetics 9: 179 Histone modifications around HBB locus Known CRMs UCSC genes trithorax Polycomb Transcription associated mark DNase hypersensitive sites Distributions at all GenCode TSSs Symmetrical distribution of: - H3K4me3, H3K4me2 - H3Ac, H4Ac, DHS - E2F1, E2F4, Myc, Pol II Birney et al. (2007) Nature 477: 799-816 Distribution of histone modifications and factor binding around regulatory regions • Promoters – H3K4me3, H3K4me2 – E2F1, E2F4, Myc, Pol II • Distal HSs – H3K4me1: enhancers – CTCF: insulators Birney et al. (2007) Nature, 447:799-816 Enhancers predicted from chromatin signatures (2009) Nature 459: 108-112 Enhancer predictions in human cells Characteristics and validation of predicted enhancers Data Resources for Genomics of Gene Regulation UCSC Genome Browser • Visualize data described in publications, e.g. – Expression data • – Regulation • • • • • • Affymetrix gene arrays, GNF, Su et al. 2004 Kim et al. 2005, PICs (TAF1) Kim et al., 2008, CTCF Boyle et al., 2008, DNase hypersensitive sites Heintzman et al., 2009, Enhancers predicted by H3K4me1 Mikkelsen et al., 2007, Chromatin modifications in pluripotent and lineage-committed cells ENCODE project, Production phase – Expression • • – Affy high density tiling arrays RNA-seq from several sources (CSHL, Helicos) Regulation • • • • • • • Broad histone modifications HAIB DNA methylation Open Chromatin UW DNase HS HAIB TFBS Yale TFBS SUNY RBP Factor occupancy and DNase hypersensitivity ENCODE Tracks: Broad histone modifications, Open chromatin, UW DHS, Yale TFBSs HS5 Locus control region 4 3 2 1 Collated sets of published regulatory regions • http://www.bx.psu.edu/~ross/dataset/Reguldata.html • Noncoding DNA segments with high regulatory potential • PRPs: Intersection of the High RP segments and the PReMods (clusters of conserved transcription factor binding site motifs) • Most constrained DNA segments, phastCons • DNase hypersensitive sites in CD4+ T cells • DNA segments occupied by CTCF in primary fibroblasts • Preinitiation complexes (TAF1) in IMR90 cells • Predicted erythroid cis-regulatory modules GeneTrack • Genomic data analysis and integration – Istvan Albert, Frank Pugh, et al., PSU – http://genetrack.bx.psu.edu/ • Install on your system • Gallery of data for visualization – Yeast H2AZ nucleosome predictions, 454 sequencing – Drosophila H2AZ nucleosome predictions, 454 sequencing Yeast nucleosome map HIS3: nucleosomefree region modENCODE http://www.modencode.org/ Worm and Fly Gene annotations Expression Chromatin modifications TFBs in vivo, etc. Experimental Tests in the Genomics of Gene Regulation GATA-1 is required for erythroid maturation Common myeloid progenitor MEP Hematopoietic stem cell G1E cells GATA-1 Myeloblast Common lymphoid progenitor G1E-ER4 cells Basophil Eosinophil Neutrophil Aria Rad, 2007 http://commons.wikimedia.org/wiki/Image:Hematopoiesis_(human)_diagram.png Monocyte, macrophage GATA1-induced changes in gene expression and occupancy genome-wide Genes induced or repressed after restoration of GATA1 Occupancy by TFs and histone modifications along a 60 Mb region High sensitivity and specificity of high throughput occupancy data High throughput occupancy matches known CRMs at Hbb locus Confirmed and novel regulatory regions for Gypa Known CRMs Gypa gene Response DHSs GATA1 TAL1 Trx: H3K4me1 Trx: H3K4me3 PcG: H3K27me3 Input DNA Induced genes have GATA1 occupied segments close to their TSS DNA segments occupied by GATA-1 were tested for enhancer activity on transfected plasmids Occupied segments Some of the DNA segments occupied by GATA1 are active as enhancers Cheng et al. (2008) Genome Research 18:1896-1905 Binding site motifs in occupied DNA segments can be deeply preserved during evolution Consensus binding site motif for GATA-1: WGATAR or YTATCW 5997 constrained 7308 not constrained 2055 no motif All GATA1-occupied segments active as enhancers are also occupied by SCL and LDB1 Genetic Determinants of Variation in Gene Expression Variation of gene expression among individuals • Levels of expression of many genes vary in humans (and other species) • Variation in expression is heritable • Determinants of variability map to discrete genomic intervals • Often multiple determinants • This variation indicates an abundance of cis-regulatory variation in the human genome • For example: – Microarray expression analyses of 3554 genes in 14 families • Morley M … Cheung VG (2004) Nature 430:743-747 - Expression analysis of about 16 HapMap individuals • Storey et al. (2007) AJHG 80: 502-509 – Expression analysis of all 270 individuals genotypes in HapMap • Stranger BE … Dermitzakis E (2007) Nature Genetics 39:1217-1224 Variation in expression between populations Figure 5.Allele-specific qPCR analysis of SH2B3. a, Log2-fold change of SH2B3 expression for all CEU and YRI individuals, relative to the average expression level in the YRI sample obtained from allele-specific qPCR. The distribution of SH2B3 expression is significantly different between samples (t-test, P= .0157), which confirms the microarray results. b, Allele-specific qPCR of a coding polymorphism (rs1107853), which demonstrates that the log2-fold change of the G allele relative to the A allele is significantly different between heterozygous DNA (Het DNA) and heterozygous cDNA (Het cDNA) samples (t-test, P= .00118). Storey et al., 2007, AJHG 80:502-509 Mapping determinants of expression variation • • Stranger et al., 2007, Nature Genetics 39:1217-1224 Expression analysis of EBV-transformed lymphoblastoid cells from all 270 individuals genotypes in HapMap – – – – • 30 Caucasian trios (90) of European descent in Utah (CEU) 30 Yoruba trios (90) from Ibadan, Nigeria (YRI) 45 unrelated Chinese individuals from Beijing Univ (CHB) 45 unrelated Japanese individuals from Tokyo (JPT) Measure levels of expression of 47,294 probes (about 24,000 genes) in each individual – Focus on 13,643 genes “selected on criteria of variance and population differentiation” • • Already know genotypes at about 2.2 million SNPs for each individual (HapMap) Test for significant association of variation at each SNP with variation in expression of each gene – Linear regression model – Spearman rank correlation test • Evaluate significance of regression P values by 10,000 permutations of the data, focus on those associations above the 0.001 permutation threshold Association of SNPs with expression • Significant association between expression and cisSNPs (within 1 Mb) • 831 genes in at least one population • 310 genes in at least 2 populations • 62 genes in all 4 populations • Also find associated SNPs in trans: perhaps regulatory proteins Stranger et al., 2007, Nature Genetics 39:1217-1224 Location of expression-associated SNPs • Most are “close” to transcription start site (TSS) • Symmetrical arrangement (similar to biochemical features of promoters) • Three of the SNPs have been shown to affect promoter activity in transfection assays (Hoogendoorn et al. (2004) Human Mutation 24: 35-42 Figure 4 Properties of significant cis associations as a function of SNP distance from the transcription start site. Stranger et al., 2007, Nature Genetics 39:1217-1224 Relevance to human health • "We predict that variants in regulatory regions make a greater contribution to complex disease than do variants that affect protein sequence” – Manolis Dermitzakis, ScienceDaily Risk loci in noncoding regions (2007) Science 316: 1336-1341 Biochemical features of DNA in CRMs Accessible to cleavage: DNase hypersensitive site Clusters of binding site motifs Bound by specific transcription factors Coactivators Pol Pol IIa II Associated with RNA polymerase and general transcription factors Nucleosomes with histone modifications: Acetylation of H3 and H4 Methylation of H3K4 Candidate functions in T2D SNP intervals Overlap of SNP rs564398 with DHS suggests a role in transcriptional regulation, but overlap with an exon of a noncoding RNA suggests a role in post-transcriptional regulation. Different hypotheses to test in future work.