* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download presentation
Survey
Document related concepts
Citric acid cycle wikipedia , lookup
Fatty acid metabolism wikipedia , lookup
Point mutation wikipedia , lookup
Fatty acid synthesis wikipedia , lookup
Enzyme inhibitor wikipedia , lookup
Nucleic acid analogue wikipedia , lookup
Genetic code wikipedia , lookup
Oligonucleotide synthesis wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Biochemistry wikipedia , lookup
Amino acid synthesis wikipedia , lookup
Biosynthesis wikipedia , lookup
Proteolysis wikipedia , lookup
Ribosomally synthesized and post-translationally modified peptides wikipedia , lookup
Transcript
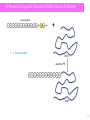
Chemo-enzymatic peptide synthesis (CEPS): a generally applicable, traceless ligation technology for the synthesis of longer peptides and peptide-to-protein conjugates. Belgian peptide group meeting February 18th, 2016 Timo Nuijens [email protected] Enzypep: The Company Founded: July 2012 as spin-off from DSM Location: Brightlands Campus in Geleen (NL) Team: Dr. Timo Nuijens Dr. Ana Toplak, B.Sc. B. Sc. Mathijs B.A.C. van de Meulenreek M.Sc. Michel Goldbach M.Sc. Marcel Schmidt Dr. Peter J.L.M. Quaedflieg (hire in basis) Funding: 2 Private investors and 3 investment funds Goals: Development of processes for “long” (pharmaceutical) peptides, cyclic peptides and peptide-to-protein conjugates with significantly lower costs (CEPS). 2 Enzypep: Initial CEPS Concept • During SPPS, yield loss increases exponentially with chain length. • Many close eluting impurities, difficult prepartive HPLC purification • Better use short fragments and segment condensation • Less and better separated impurities Full SPPS Yield as Function of Chain Length and Coupling Efficiency 100.0% Crude Yield 80.0% 99.5% 60.0% 99.0% 40.0% 98.0% Fragment approach 97.0% 20.0% 95.0% 90.0% 0.0% 0 5 10 15 20 25 30 35 40 Number of Amino Acids 3 Fragment condensation: SPPS-LPPS and NCL Chemical Hybrid (SPPS-LPPS) Fragment Condensation • racemization except sequences with C-terminal Gly or Pro • fully protected peptides have poor solubility and are difficult to purify Other chemical ligation techniques (such as NCL or KAHA) • sequence dependent • NCL, thioester instability 4 Fragment condensation: CEPS Chemo-enzymatic Peptide Synthesis (CEPS) in Aqueous Solution • no racemization • no side-chain protection = better solubility, easier purification • selective, does not recognize D-amino acids • no side-reactions as encountered using chemical fragment condensation Potential drawbacks: • C-terminal ester needed (can be expensive/unstable) • hydrolytic side-reactions: synthesis to hydrolysis ratio (S/H ratio) • tight sequence specificity (not generally applicable) • enzyme instability 5 Chemo-enzymatic Peptide Synthesis (CEPS) Most often used: proteases How can hydrolytic side-reactions (intrinsic hydrolase activity) be minimized? • Reaction optimization • Omit water • Special esters, substrate mimetics (Bordusa et al.) • Natural occurring ligases: Sortases Inteins Butelase Aqueous media Require specific amino acid motifs May not be traceless Peptides tuned to requirements of enzymes • Designed ligases (mutagenesis): Peptiligases Enzymes tuned to requirements of peptides 6 Ligases derived from nature Sortase Reaction Not traceless sortase footprint Butelase Reaction butelase footprint Butelase 1 • Sortases, butelase and inteins for a thioester intermediate • Transpeptidation able to cleave a peptide amide bond 7 Subtiligase, designed ligases Ligases by mutagenesis, protein design - Streptoligase (Elloitt et al.) - Subtiligase (Wells et al.) Subtilisin (serine protease) • • • • S221C mutation at active site thioester intermediate P225A mutation to prevent steric crowding “double mutant” called subtiligase can be used to • • • • ligate peptide fragments cyclize peptides >12 amino acids couple peptides to proteins synthesize thioacids and thioesters Limitations: • • average yield of 50% using 10 equivalents of one fragment Subtiligase is labile, is Ca2+ dependent, difficult to produce and purify Distinct need for more stable and efficient enzymes with higher S/H ratios 8 Peptiligase Starting from a hyperstable and cation (calcium) independent variant of subtilisin (Bryan et al.) that was - stable to additives (urea and guanidiniumchloride) - stable to organic co-solvents (DMF and DMSO, up to 50 vol%) - thermostable (unfolding temperature = 70⁰C) - very hydrolytically active we incorporated the S221C and P225A mutations into this enzyme scaffold. Surprisingly, this new enzyme “peptiligase” exhibited • • • 2-fold higher S/H ratio very fast ligation reaction retention of stability towards detergents, co-solvents and temperature Furthermore • peptiligase was easy to produce and purify • little enzyme was needed (~1 mg enzyme/gram of 20-mer peptide) • perfect scaffold for further protein engineering 9 Peptiligase condensation • Peptide Cam-esters synthesized via SPPS (Nuijens et al.) Leu-Wang Resin, coupling of iodoacetic acid, Fmoc-amino acid and DiPEA • Usually side-chain unprotected peptides in buffer (pH 8) at 20⁰C. • Ligation yield usually > 90% synthesis and < 10% hydrolysis for a 10-mer + 10-mer coupling using only 1.1 equivalents of the amine or Cam-ester fragment 10 Peptiligase Specificity medium medium Limitations: very tight amine fragment substrate profile, preference for Gly, Ala and Ser Action: broaden peptiligase specificity, S/H ratio, activity via site-directed mutagenesis Started more than 4 years ago Peptiligase Specificity :P’1 pocket HPLC product yield* Development of 1st, 2nd and 3rd generation mutants: P'1 pocket • Improvement of the P'1 pocket specificity • All amino acids in P’1 pocket except Pro are now well accepted *Suboptimal conditions to demonstrate differences 12 Peptiligase Specificity: P’2 Pocket Development of 1st, 2nd and 3rd generation mutants: P‘2 pocket 90% 80% • Improvement of the P‘2 pocket specificity • >80% of the amino acids in P’2 pocket are now well accepted • In development: “Omniligase”, a peptiligase that can couple almost all sequences. Commercially available (Iris) HPLC product yield* 70% 60% Ptl 50% 1st 40% 2nd 30% 3rd 20% 10% 0% *Suboptimal conditions to demonstrate differences 13 Peptiligase Specificity: Selectivity Development of 1st, 2nd and 3rd generation mutants: P'1 pocket 90.0% • Also highly specific enzymes -Large/small -Polar/hydrophobic -Charge • For both P’1 and P’2 HPLC product yield* 80.0% 70.0% 60.0% 50.0% Sel-1 40.0% Sel-2 • 30.0% 20.0% 10.0% • 0.0% Often no N- or Cterminal protecting groups are needed >10,000 unique (specific) ligases now at our disposal *Suboptimal conditions to demonstrate differences 14 S/H Ratio in 1st, 2nd & 3rd Generation Mutants S:H Ratio S/H Ratio for 1st, 2nd and 3rd generation Peptiligase mutants 9 8 7 6 5 4 3 2 1 0 S/H Ratio • Besides improving the substrate scope, the S/H ratio is also much higher • An S/H of 9 means 90% synthesis and 10% Camester hydrolysis • We performed 10 + 10 coupling reactions with S/H ratio’s of >50, corresponding to 98% synthesis and only 2% hydrolysis • No other side reactions as encountered using chemical fragment condensation 15 Example 1: Exenatide (2 Fragment Approach) Exenatide has no Gly or Pro residues at strategic positions and is therefore most often synthesized via full SPPS. Typical yield of commercial processes = 20 – 25%. 1-21 22-39 Cam-ester Fragment (unprotected): H-His1-Gly2-Glu3-Gly4-Thr5-Phe6-Thr7-Ser8-Asp9-Leu10-Ser11-Lys12-Gln13-Met14Glu15-Glu16-Glu17-Ala18-Val19-Arg20-Leu21-OCam-Leu-OH Amine Fragment (unprotected): H-Phe22-Ile23-Glu24-Trp25-Leu26-Lys27-Asn28-Gly29-Gly30-Pro31-Ser32-Ser33-Gly34Ala35-Pro36-Pro37-Pro38-Ser39-NH2 Coupling: 2nd generation peptiligase Amine Fragment, using Rink resin: Crude fragment: 70.6% purity, 65% yield; Single purification: 96.0% purity, 52% yield Cam-ester Fragment, using CTC resin Crude fragment: 72.5% purity, 69% yield, Single purification: 97.1% purity, 56% yield 16 CEPS of Exenatide (2-Fragment Strategy) • • • • • • • 2nd generation peptiligase At gram scale No protection at all, API in one step 1.1 equivalent amine fragment Conversion to product 87% 30 g/L Exenatide product Crude reaction mixture for prep HPLC 17 CEPS of Exenatide (2-Fragment Strategy) Exenatide (Pure Fragments) Scale: Crude peptide: Single purification: Overall yield: 1.0 g, 3.0 wt%, amine excess = 1.1 equiv. 78.9% purity, 87% yield 99.6% purity, 78% yield (based on Cam-ester) 43.5% (based on resin loading of Cam-ester) NaClO4-ACN • • • • TFA-ACN Overall yield 2-fold to better than straight-through SPPS Purity exceeds innovator’s specifications Chiral integrity confirmed by C.A.T. Cost-price reduction estimated around 40% - 50% 18 CEPS What more can we do more with our ligases ?? • Head-to-tail cyclisation • N- to C- polymerisation • Maximum ligation length ? • Peptide to protein conjugation • Peptide to polymer • Etc, etc 19 Macrocylic Peptides: Exenatide, 39-mer H-Phe22-Ile23-Glu24-Trp25-Leu26-Lys27-Asn28-Gly29-Gly30-Pro31-Ser32-Ser33-Gly34Ala35-Pro36-Pro37-Pro38-Ser39-His1-Gly2-Glu3-Gly4-Thr5-Phe6-Thr7-Ser8-Asp9-Leu10Ser11-Lys12-Gln13-Met14-Glu15-Glu16-Glu17-Ala18-Val19-Arg20-Leu21-OCam-Leu-OH Parameter Conditions Enzyme Buffer Temperature Time Conversion Broad specificity Ptl Phosphate buffer, pH 8.3 20°C 5 min 98% 1 39 N to C coupling point • Efficiently cyclised many more peptides ranging fom 12-50 amino acids 20 N-Terminal Conjugation: Exenatide to Serum Albumin + 1 + 1 Condensation selective Ptl 21 N-Terminal Conjugation: Peptide to XTEN(144) and XTEN(864) model peptide + 1 + 1 Condensation selective Ptl 22 Conclusions • Peptiligase (extremely stable ligase) • Efficient condensation of unprotected peptide segments in water • 1st, 2nd and 3rd generation variants: -”Omniligase”, can couple almost all peptide sequences -”Sequence-specific ligases”, very selective • Synthesis of Exenatide • Head-to-tail cyclisation • Peptide-to-protein or peptide to polymer conjugation 23 Acknowledgements University of Groningen: Prof Dick B. Janssen Dr. Ana Toplak Dr. Bian Wu (now at Enzypep) Enzypep: Dr. Peter J.L.M. Quaedflieg B.Sc. Mathijs B.A.C. van de Meulenreek M.Sc. Marcel Schmidt M.Sc. Michel Goldbach M.Sc. Francesco Ventura Scientific advisory board: Prof Dick B. Janssen Dr. Paul ten Kortenaar Dr. Rodney Lax Dr. Roland Callans Dr. Rinus Broxterman (University of Groningen) (MSD, Diosynth) (PolyPeptide Group) (Corden) (DSM) 24