* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download Summary of Chapter 24
Basal metabolic rate wikipedia , lookup
Oligonucleotide synthesis wikipedia , lookup
Ribosomally synthesized and post-translationally modified peptides wikipedia , lookup
Microbial metabolism wikipedia , lookup
Point mutation wikipedia , lookup
Proteolysis wikipedia , lookup
Nucleic acid analogue wikipedia , lookup
Butyric acid wikipedia , lookup
Oxidative phosphorylation wikipedia , lookup
Nitrogen cycle wikipedia , lookup
Adenosine triphosphate wikipedia , lookup
Evolution of metal ions in biological systems wikipedia , lookup
Protein structure prediction wikipedia , lookup
Metalloprotein wikipedia , lookup
Peptide synthesis wikipedia , lookup
Fatty acid metabolism wikipedia , lookup
Glyceroneogenesis wikipedia , lookup
Fatty acid synthesis wikipedia , lookup
Genetic code wikipedia , lookup
Citric acid cycle wikipedia , lookup
Biosynthesis wikipedia , lookup
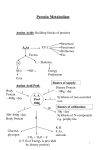
Takusagawa’s Note Chapter 20 (Summary) Summary of Chapter 20 1. Transamination • Major problem in amino acid degradation is elimination of -NH2 since NH3 from -NH2 is very toxic. • There are two ways to remove -NH2 from amino acids. 1. Transamination: R-NH2 → Glu-NH2 → Asp-NH2 →→ Urea + → α-ketoglutarate + NH →→ Urea 2. Oxidative deamination: R-NH → Glu-NH NAD 2 2 3 • Transaminases contain co-enzyme, pridoxal-5’-phosphate (PLP), which is covalently attached to the Lys residue of the enzyme via Schiff base linkage (E-Lys-PLP). • Aminotransferase reaction: AA1 ↔ α-Keto acid1 1. Transimination: E-Lys-PLP + AA1 ↔ [E-PLP-AA1] ↔ E-Lys + PLP-AA1 2. Tautomerization: PLP-AA1 ↔ α-Keto acid1-PMP [PMP = PLP-NH2] 3. Hydrolysis: α-Keto acid1-PMP + H2O ↔ α-Keto acid1 + PMP • Reverse aminotransferase reaction: α-Keto acid2 ↔ AA2 3’. α-Keto acid2 + PMP ↔ α-Keto acid2-PMP 2’. α-Keto acid2-PMP ↔ PLP-AA2 1’. PLP-AA2 + E-Lys ↔ [E-PLP-AA2] ↔ E-Lys-PLP + AA2 • Ammonia is expeled as urea (amminals) and uric acid (birds). 2. Urea cycle • Urea is formed from 1. Ammonia (NH3) from oxidative deamination 2. Amine (-NH2) of Asp 3. Bicarbonate (HCO3-) • Reactions 1. Formation of carbamoyl phosphate by carbamoyl phosphate synthetase. HCO3- + NH3 + 2ATP → H2N-CO(OPO32-) + 2ADP + Pi 2. Condensation of ornitine and carbamoyl to form citrulline by ornithine transcarbamoylase. Ornitine + H2N-CO(OPO32-) → Citrulline + Pi 3. Condensation of citrulline and Asp to form argininosuccinate by argininosuccinate synthetase Citrulline + Asp + ATP → Argininosuccinate + AMP + PPi 4. Cleavage of argininosuccinate to Arg and fumarate by argininosuccinase Argininosuccinate → Arg + Fumarate 5. Hydrolysis of Arg to urea and ornitine by arginase Arg + H2O → Urea + Ornitine • Overall reaction uses 4 “high energy” phosphate bond hydrolysis. CO2 + NH3 + Asp + 2H2O + 3ATP → Urea + Fumarate + 2ADP + AMP + 2Pi + PPi (→ 2Pi) • Oxidation of urea cycle produces 2NADH (= 6ATP). • Krebs bicycle: Urea cycle and aspartate-argininosuccinate shunt of citric acid cycle. • Urea cycle is regulated by [N-acetyl-glutamate] which activates carbamoyl phosphate synthetase. 1. Breakdown of proteins produces amino acids including Glu. 2. Glu is acetylated by acetyl-glutamate synthase. 3. N-acetyl-glutamate activates urea cycle. 1 Takusagawa’s Note Chapter 20 (Summary) • Ammonia produced in all tissues are transported by Gln and Ala. 1. NH4+ + Glu + ATP → Gln + ADP + Pi + H+ in tissue + + in liver Gln + H2O → Glu + NH4 ; NH4 → Urea 2. Glucose-alanine cycle: Glucose → Pyruvate + Glu → Ala + α-Ketoglutarate in Tissue Ala + α-Ketoglutarate → Pyruvate + Glu in Liver Glu → NH3 → Urea Pyruvate → Glucose 2. Amino acid’s skeleton metabolism • 20 amino acids are converted to 7 common intermediates. Those are: Both glucogenic and ketogenic intermediate 1. Pyruvate Ala, Cys, Gly, Ser, Thr, Tyr 2. α-ketoglutarate Glucogenic intermediates (form glucose) 3. Succinyl-CoA Arg, Glu, Gln, His, Pro, Ile, Met, Val, Asp, 4. Fumarate Phe, Tyr, Asn 5. Oxaloacetate 6. Acetyl-CoA 7. Acetoacetate Ketogenic intermediates (form ketone bodies) Ile, Leu, Thr, Lys, Phe, Trp, Tyr 3. Metabolism of C1 unit (Tetrahydrofolate cofactors) • THF functions to transfer C1 unit in several oxidation states. • C1 units carried by THF are: Methyl (-CH3); Methylene (-CH2-); Formyl (-CH=O); Formimino (-CH=NH); Methenyl (-CH=) • C1 units are donated from Ser, Gly, HCO3-, His to THF. • Sulfonamides competitively inhibit bacterial synthesis of THF, because those structures are analogues to p-aminobenzoic acid moiety of THF. 4. Synthesis of some amino acids. • Ala, Asn, Asp, Cys, Glu, Gln, Gly, Pro, Ser, Tyr are nonessential amino acids. • Glycerate-3-phosphate →→→ Ser → Gly → Cys • Pyruvate → Ala • α-Ketoglutarate → Glu → Gln; Glu→→→Pro • Oxaloacetate → Asp → Asn THF→5,10-methylene THF → Gly • Ser • Phe → Tyr by phenylalanine-4-monooxygenase. • Glutamine synthetase is a central control point in the nitrogen metabolism, since Gln is an amino group donor and a storage of ammonia. 1. Feedback inhibition: His, Tyr, carbamoyl phosphate, AMP, CTP, glucosamine-6phosphate (all end products from Gln) are allosteric inhibitors. Ala, Ser and Gly also inhibit since high concentration of these amino acids is a signal of saturation of the citric acid cycle . 2. Covalent modification (adenylylation-deadenylylation and uridylylation-deuridylylation) • Inhibition under conditions of nitrogen excess. 2 Takusagawa’s Note Chapter 20 (Summary) 5. • • 6. • • 7. • 1. High [Gln] activates uridylyl-removing enzyme in uridylyltransferase. 2. Uridylyl-removing enzyme removes UMP from adenylyltransferase. 3. Adenylyltransferase inactivates glutamine synthetase by adenylylation. • Activation under conditions of nitrogen limitation. 1. High [α-ketoglutarate] activates uridylyltransferase. 2. Uridylyltransferase uridylylates adenylyltransferase. 3. Uridylylated adenylyltransferase activates glutamine synthetase by removes AMP. S-Adenosylmethionine (SAM) is synthesized from Met and ATP. is the major methyl group donor Porphyrin synthesis Precursors of porphyrin biosynthesis are Gly and succinyl-CoA. Enzyme contains 1. Gly + Succinyl-CoA → δ-Aminolevlinate (ALA) + CO2 + CoA PLP. 2. 2ALA → Porphobilinogen (PBG) 3. 4PBG → Hydroxymethylbilane → Uroporphyrinogen III → Various hemes Overall heme biosynthesis is taken place in both cytosol and mitochondrion. Various amine biosynthesis Some of amines are important neurotransmitters, hormones, and reducing agents. Glu → γ-Aminobutyric acid (GABA) His → Histamine Trp → Serotonin Tyr → Dihydroxyphenylalanine (L-DOPA) → Dopamine → Norepinephrine → Epinephrine Glu + Cys + Gly → Glutathione (GSH) 3